Introduction
The genus Vibrio stands as a testament to bacterial diversity, a sprawling collection of Gram-negative bacteria – approximately 139 species – thriving in marine environments across the globe [Thompson et al., 2004]. Since its initial characterization with the isolation of Vibrio cholerae by Filippo Pacini in 1854 [Pacini, 1854], Vibrio has been a subject of intense scientific scrutiny. This is partly due to the genus’s cultivability and the clinical relevance of at least a dozen species known to cause human disease [Chakraborty et al., 1997]. The notoriety of V. cholerae and V. parahaemolyticus is amplified by their association with pandemic outbreaks [Karaolis et al., 1995; Matsumoto et al., 2000], while V. vulnificus is recognized for its potential to induce severe sepsis, particularly in individuals with compromised immune systems [Strom and Paranjpye, 2000]. Beyond human health, Vibrio species impact commercially vital aquaculture, with species like V. harveyi and V. anguillarum posing threats to shrimp and fish farming, respectively [Austin and Zhang, 2006; Frans et al., 2011], and V. coralliilyticus and V. shiloi acting as coral pathogens [Rosenberg et al., 2007]. Despite these pathogenic roles, many Vibrio species are benign, and the genus itself is considered “conditionally rare” in many environments [Shade et al., 2014; Thompson and Polz, 2006; Thompson et al., 2006].
The ecological distribution of Vibrio is remarkably broad, spanning riverine, estuarine, and marine ecosystems worldwide [Thompson et al., 2004]. Sea surface temperature can act as a limiting factor, often restricting growth to warmer latitudes due to the mesophilic nature of many species [Takemura et al., 2014]. However, the discovery of Vibrio diabolicus (strain CNCM I-1629) from a polychaete at a deep-sea hydrothermal vent in the East Pacific Rise [Raguénès et al., 1997] and V. antiquarius (strain EX25) from seawater near a sulfide chimney in the same region [Hasan et al., 2015] challenged this coastal-centric view. These deep-sea isolates suggested a broader ecological niche for the genus, extending far beyond typical coastal habitats.
Early research suggested that V. diabolicus CNCM I-1629 and V. antiquarius EX25 possessed unique genetic adaptations for survival in hydrothermal vent ecosystems. For instance, the acidic exopolysaccharide (EPS) produced by V. diabolicus CNCM I-1629 was hypothesized to provide protection against the high concentrations of metallic sulfides prevalent in sulfide chimney environments [Raguénès et al., 1997; Goudenège et al., 2014]. Similarly, the genome of V. antiquarius EX25 revealed an alkyl hydroperoxide reductase, an enzyme also found in the deep-sea tubeworm endosymbiont Riftia pachyptila, suggesting a role in scavenging endogenous hydrogen peroxide in the deep-sea environment [Hasan et al., 2015; Markert et al., 2007]. Intriguingly, both species also carried genes for hemolysins and type III secretion systems (T3SS), typically associated with virulence in pathogenic Vibrio species [Goudenège et al., 2014; Hasan et al., 2015]. The persistence of these virulence-associated factors in deep-sea species raised questions about their broader ecological roles, potentially extending beyond pathogenicity to environmental fitness [Hasan et al., 2015].
The global distribution, diversity, and potential virulence of these deep-sea Vibrio species remained largely unexplored. The hypothesis that V. diabolicus and V. antiquarius might have a cosmopolitan distribution gained traction with the known adaptability of related Vibrio species like V. alginolyticus and V. parahaemolyticus, recognized as opportunitrophs capable of exploiting diverse and ephemeral resources [Polz et al., 2006]. The detection of V. antiquarius EX25 open reading frames (ORFs) in numerous metagenomic datasets [Hasan et al., 2015] and the isolation of multiple V. diabolicus strains from a mid-Atlantic estuary [Klein et al., 2014] further supported the idea of a wider distribution for these deep-sea Vibrio.
During an extensive investigation into V. parahaemolyticus [Johnson et al., 2012; Paranjpye et al., 2012; Turner et al., 2013, 2016], Vibrio sp. 939 was isolated from Pacific oysters (Crassostrea gigas) harvested from Puget Sound in the Pacific Northwest of the United States. The initial identification of Vibrio sp. 939 and its subsequent genome sequencing prompted a detailed taxonomic analysis, involving 49 Harveyi clade genomes, to clarify its identity. This analysis revealed a compelling case of synonymy: Vibrio sp. 939, V. diabolicus Art-Gut C1 and CNCM I-1629, V. antiquarius EX25, and surprisingly, four V. alginolyticus strains (E0666, FF273, TS13, and V2) were found to be synonymous. This discovery significantly broadens the V. diabolicus species definition and provides six additional genomes for in-depth comparative genomic studies. The expanded V. diabolicus species exhibits a global distribution, evidenced by its isolation from diverse sources (horse mackerel, seawater, sediment, dentex, oyster, artemia, and polychaete) across various geographic origins (China, India, Greece, United States, East Pacific Rise, and Chile). A subsequent comparative genomic analysis of this newly defined eight-genome subclade uncovered substantial genome plasticity and a rich array of genes associated with virulence and defense. These findings necessitate a significant revision of the taxonomic understanding of V. diabolicus and V. antiquarius, previously considered distinct species. This initial exploration of the expanded V. diabolicus subclade suggests that its distribution and diversity mirror those of other Harveyi clade species, renowned for their widespread presence and ecological versatility. In essence, this research highlights how synonyms in bacterial taxonomy, much like Synonyms For Comparative terms in language, can reflect different labels for fundamentally the same entity, and how genomic comparisons are crucial for resolving these taxonomic ambiguities.
Materials and Methods
Sample Collection
Vibrio sp. 939 was isolated from Pacific oysters (C. gigas) sourced from Hood Canal, a part of Puget Sound in Washington State, USA. This isolate was among a large collection obtained by the NOAA Northwest Fisheries Science Center during a comprehensive V. parahaemolyticus investigation [Johnson et al., 2012; Paranjpye et al., 2012; Turner et al., 2013, 2016]. Oysters were meticulously scrubbed, shucked, and homogenized. Presumptive Vibrio species were then isolated through direct plating onto thiosulfate-citrate-bile salts-sucrose (TCBS) agar (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) and incubated overnight at 30°C. Vibrio sp. 939 was initially identified as V. parahaemolyticus based on the detection of the thermolabile hemolysin (tlh) gene, a marker associated with this species [Bej et al., 1999]. However, it was excluded from a previous multilocus sequence typing (MLST) study focused on V. parahaemolyticus [Turner et al., 2013] due to amplification challenges with several MLST loci. Furthermore, recent studies indicated that the V. parahaemolyticus-specific tlh primers could also amplify tlh gene variants in closely related species [Klein et al., 2014; Yáñez et al., 2015], casting doubt on the preliminary assignment.
Culture Conditions
Starting from a frozen stock culture of Vibrio sp. 939 (25% glycerol, v/v, -80°C), the isolate was streaked for isolation on lysogeny broth (LB) (Fisher Scientific, Fair Lawn, NJ, United States) supplemented with 1.5% Bacto agar (Becton, Dickinson and Company, Franklin Lakes, NJ, United States). The plates were incubated overnight (18 h) at 30°C. A single, well-isolated colony was then transferred to 5 mL of LB in a 15 mL BD Falcon tube (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) and cultured overnight (18 h) at 30°C with shaking (120 rpm) using an Excella E24 shaking incubator (Eppendorf, Hamburg, Germany).
DNA Isolation
For DNA extraction, 1 mL of the overnight bacterial culture was centrifuged at 9,400 × g for 5 minutes in an Eppendorf 5424E centrifuge (Hamburg, Germany) to pellet the cells. The cell pellet was washed twice with an equal volume of phosphate-buffered saline (PBS). Genomic DNA was then isolated from the washed cells using a ChargeSwitch gDNA Mini Bacteria Kit (Invitrogen, Carlsbad, CA, United States), following the manufacturer’s recommended protocols. The extracted DNA was quantified and its quality (A260/A280 ratio) was assessed using a BioPhotometer D30 (Eppendorf, Hamburg, Germany). The purified DNA was stored at -20°C until further use.
Genome Sequencing
Genome sequencing of the extracted DNA was performed using an Illumina MiSeq instrument at the New York University Genome Technology Center, employing paired-end sequencing chemistry (2 × 300 bp). A PCR-free version of the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, United States) was used to prepare the sequencing library. Overlapping paired reads were merged using FLASH version 1.2.11 [Magoc and Salzberg, 2011]. Adapter sequences and low-quality bases were trimmed from the merged reads using Trim Galore! version 0.4.4, a wrapper script incorporating Cutadapt [Martin, 2011] and FastQC [Andrews, 2010]. The optimal k-mer size for genome assembly was estimated using KmerGenie version 1.7 [Chikhi and Medvedev, 2014]. De novo genome assembly was then carried out using Velvet version 1.2.10 [Zerbino and Birney, 2008]. Initial genome annotation and inspection were performed using the web-based RAST annotation service and SEED Viewer [Aziz et al., 2008; Overbeek et al., 2014]. Final genome annotation was completed using the National Center for Biotechnology Information’s (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) [Klimke et al., 2009].
Phylogenetics
To determine the phylogenetic relationships of Vibrio sp. 939 with 48 closely related Harveyi clade species, including V. diabolicus, V. antiquarius, V. parahaemolyticus, V. alginolyticus, and V. harveyi (Supplementary Table S1), a maximum-likelihood phylogenetic tree was constructed. This tree was based on a set of single-copy homologous genes present in all 49 Harveyi clade genomes. The genome sequences were downloaded from NCBI. Single-copy homologous genes were identified and clustered using get_homologues (options -M -t 49 -r EX25.gbk -e) [Contreras-Moreira and Vinuesa, 2013]. These options specified the use of the OrthoMCL algorithm [Li et al., 2003] with a default E-value cutoff of 1e-05 for homolog searching, restricting clustering to single-copy homologs present in all 49 genomes, using the closed genome of V. antiquarius EX25 as a reference, and excluding clusters containing inparalogs. Homologous clusters were individually aligned using MUSCLE version 3.6 [Edgar, 2004], and alignments were trimmed to the length of the shortest sequence using trimAl version 1.4.15 [Capella-Gutierrez et al., 2009]. Trimmed alignments shorter than 250 bp were removed. The remaining trimmed alignments were concatenated using the fasta manipulation tool in Galaxy [Giardine et al., 2005]. A maximum-likelihood tree was then constructed using IQ-TREE version 1.5.5 [Nguyen et al., 2014] with 1,000 ultrafast bootstraps [Minh et al., 2013] and the best-fit model (GTR+I+G4) as determined by ModelFinder [Kalyaanamoorthy et al., 2017]. The resulting phylogenetic tree was annotated using FigTree version 1.4.3.
Phylogenetic Network
To visualize potential conflicting phylogenetic signals resulting from reticulate evolutionary processes, a Neighbor-Net phylogenetic network [Bryant and Moulton, 2004] was constructed using the same concatenated multilocus alignment as used for the phylogenetic tree. The network was generated using SplitsTree4 version 4.14.4 [Huson and Bryant, 2006] with default settings and 1,000 bootstraps.
Genome Similarity
To quantify genome similarity, pairwise average nucleotide identity (ANI) values were calculated between the genomes of V. diabolicus Art-Gut C1 and CNCM I-1629, Vibrio sp. 939, V. antiquarius EX25, and V. alginolyticus 12G01, 40B, E0666, FF273, K01M1, NBRC 15630, TS13, and V2. These comparisons were performed using JSpeciesWS [Richter et al., 2016], a web server implementation of the JSpecies Taxonomic Thresholds Program [Richter and Rosselló-Móra, 2009]. JSpeciesWS calculates ANI values between a query and a reference genome, with a 95–96% similarity threshold generally accepted for prokaryotic species delineation. To visually represent the sequence similarity among the eight V. diabolicus subclade genomes (defined in this study as V. diabolicus Art-Gut C1 and CNCM I-1629, Vibrio sp. 939, V. antiquarius EX25, and V. alginolyticus E0666, FF273, TS13, and V2), a circular blast map was generated using the BLAST Ring Image Generator (BRIG) [Alikhan et al., 2011]. For this map, the lower and upper identity thresholds were set to 92% and 96%, respectively, and the closed genome of V. antiquarius EX25 served as the reference genome. V. alginolyticus NBRC 15630 genome was included for contrast. Genome attributes, such as genome size, %GC content, total genes, protein-coding genes, and RNA genes, were also calculated to facilitate comparison of these eight genomes.
Pangenome Analysis
Pangenome analysis was performed to assess the genomic diversity within the V. diabolicus subclade. Two methods were employed to determine the pangenome of the eight V. diabolicus subclade genomes. First, the pangenome was estimated using get_homologues with the option t = 0 to report all homologous gene clusters. The accompanying parse_pangenome_matrix.pl script was used to identify core genes (present in all eight strains) and accessory genes (present in fewer than eight strains). Recognizing that get_homologues excludes clusters with fewer than three sequences and non-coding sequences, a second pangenome analysis was conducted using Panseq [Laing et al., 2010] with default parameters. Panseq estimates the pangenome through all-versus-all alignment of genomic regions and reports core, accessory, and pangenome sizes. For this analysis, the core genome was defined as genomic regions present in all eight genomes. To identify singletons (genomic regions unique to one genome), Panseq’s Novel Region Finder was used. Novel regions were then filtered by length (10 Kb cutoff) to identify probable genomic islands (GIs). These GIs were further examined using the web-based SEED Viewer (blastn E-value cutoff 1e-05) [Overbeek et al., 2014] to identify genes potentially linked to habitat-specific survival or persistence.
Subclade-Associated Genes
To identify genes specifically associated with the V. diabolicus subclade, the output from the get_homologues pangenome analysis was parsed using the parse_pangenome_matrix.pl script. This analysis aimed to identify genes present in all eight V. diabolicus subclade strains but absent from the other 41 strains. Subclade-associated genes and their protein products were individually aligned using MUSCLE version 3.6 [Edgar, 2004]. The average sequence identity for each alignment was calculated using trimAl version 1.4.15 [Capella-Gutierrez et al., 2009].
Metagenome Survey
To investigate the environmental prevalence of V. diabolicus, a metagenome survey was performed. Protein sequences encoded by the most conserved non-hypothetical subclade-associated genes were used as queries against the NCBI Metagenome Protein Database (env_nr) using default blastp parameters. Database hits were considered significant if the sequence identity was comparable to the subclade protein alignments (greater than 95%).
Potential Virulence
The presence and diversity of genes related to virulence and defense were investigated using SEED subsystem feature counts in the web-based SEED Viewer. The eight genomes of the V. diabolicus subclade were screened for proteins with significant matches (blastp with E-value cutoff 1e-05) to five SEED subsystems: (1) Adhesion, (2) Toxins and Superantigens, (3) Bacteriocins and Ribosomally Synthesized Antibacterial Peptides, (4) Resistance to Antibiotics and Toxic Compounds, and (5) Invasion and Intracellular Resistance. Specifically, β-lactamase proteins, enzymes conferring resistance to β-lactam antibiotics, were analyzed. β-lactamase proteins were aligned using MUSCLE version 3.6 [Edgar, 2004], trimmed using trimAl version 1.4.15 [Capella-Gutierrez et al., 2009], and a maximum-likelihood tree was constructed using IQ-TREE version 1.5.5 [Nguyen et al., 2014] with 1,000 ultrafast bootstraps [Minh et al., 2013] and the best-fit model (WAG) determined by ModelFinder [Kalyaanamoorthy et al., 2017]. Additionally, β-lactamase proteins (present in all genomes) and fosfomycin resistance proteins (V. alginolyticus TS13 and V2) were searched against the Antibiotic Resistance Genes Database (ARDB) using default parameters (blastp with E-value cutoff 1e-05) [Liu and Pop, 2009].
Antibiotic Susceptibility
β-lactamase production was assessed using Cefinase disks (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) following the manufacturer’s instructions. Susceptibility to selected β-lactam antibiotics was determined as previously described [Letchumanan et al., 2015]. V. antiquarius 939 was grown overnight (18 h) on Mueller-Hinton agar plates (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) supplemented with 1.5% NaCl at 37°C. Overnight growth was resuspended in filter-sterilized 0.85% NaCl solution, adjusted to a 0.5 McFarland standard, and fresh Mueller-Hinton agar plates were seeded with a bacterial lawn using a sterile cotton swab. Antibiotic disks containing penicillin (10 units), ampicillin (10 μg), cephalothin (30 μg), and carbenicillin (100 μg) (Becton, Dickinson and Company, Franklin Lakes, NJ, United States) were placed on the lawn, and plates were incubated overnight (18 h) at 37°C. Zones of inhibition were measured and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [Clinical and Laboratory Standards Institute [CLSI], 2010]. The pandemic type strain of V. parahaemolyticus (RIMD2210633) [Nasu et al., 2000] was tested for comparative purposes.
Results
Genome Sequencing
The draft genome of Vibrio sp. 939 consisted of 46 contigs, totaling 5,430,661 bp in length, with a 44.6% GC content. The N50 of the assembly was 855,346 bp, and contig lengths ranged from 570 bp to 1,035,854 bp. PGAP annotation identified 5,049 genes, including 4,876 protein-coding genes, 112 RNA genes (13 rRNA, 95 tRNA, and 4 other RNA), and 61 pseudogenes. Based on initial evaluation using the SEED Viewer’s “View Closest Neighbors” feature, the whole-genome shotgun project was deposited as V. antiquarius 939 at DDBJ/ENA/GenBank under accession number AOJB00000000.
Phylogenetic Tree
To construct a phylogeny of the Harveyi clade, a homologous gene search identified 1,125 gene clusters shared among all 49 genomes. After applying a 250 bp length filter, 16 clusters were removed, resulting in 1,109 single-copy homologs. Concatenation of these homologs yielded a multilocus alignment of 1,034,083 bp, comprising 595,014 constant sites, 404,946 parsimony-informative sites, and 217,816 distinct site patterns. ModelFinder determined GTR+I+G4 as the best-fit model according to Bayesian information criterion (BIC) scores and weights.
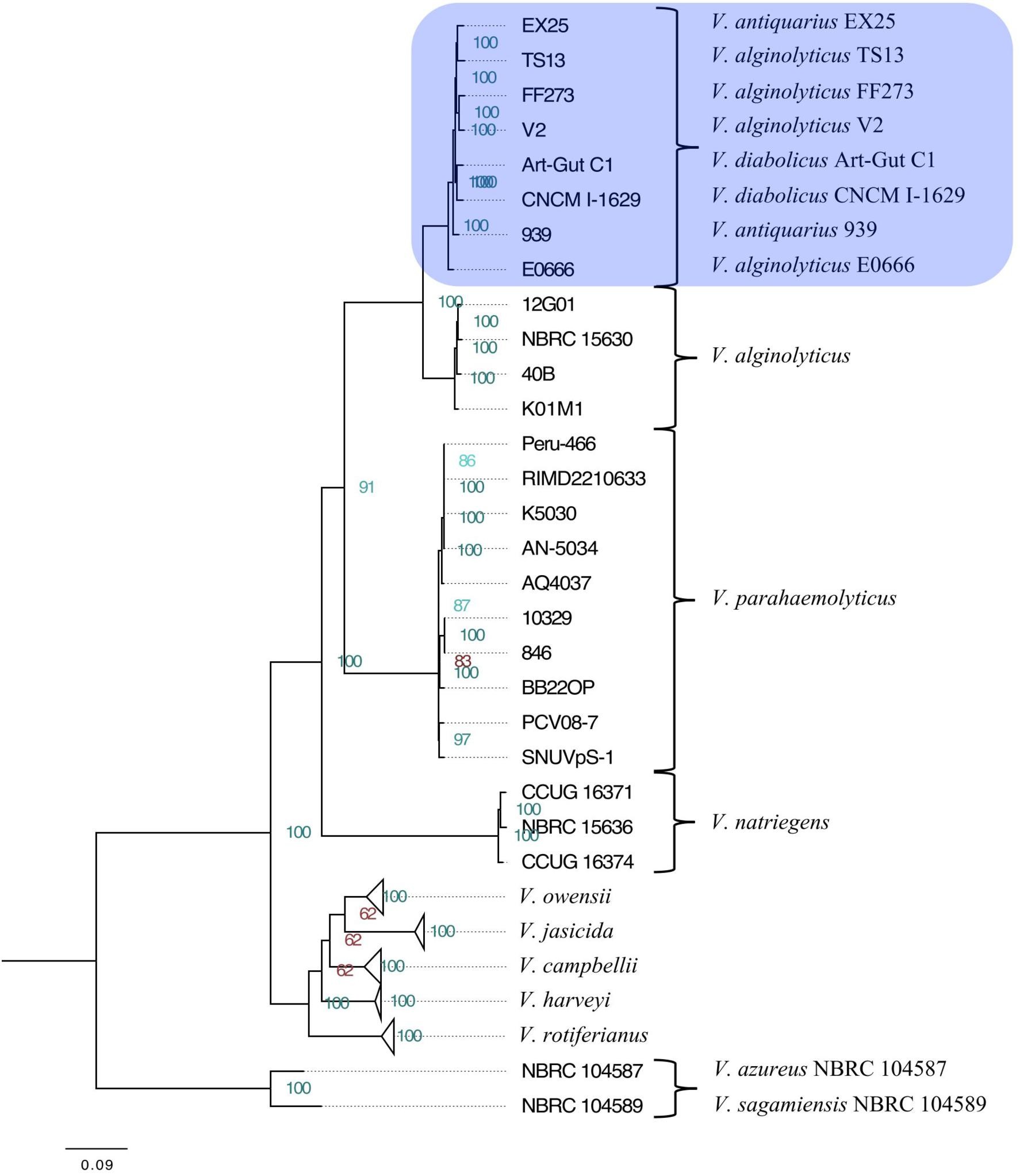
The maximum-likelihood phylogeny (Figure 1) resolved the Harveyi clade into eleven subclades: V. alginolyticus, V. azureus, V. campbellii, V. harveyi, V. jasicida, V. natriegens, V. owensii, V. parahaemolyticus, V. rotiferianus, V. sagamiensis, and a mixed subclade. This mixed subclade was composed of two V. diabolicus strains (Art-Gut C1 and CNCM I-1629), two V. antiquarius strains (939 and EX25), and four V. alginolyticus strains (E0666, FF273, TS13, and V2). The complete, uncollapsed phylogenetic tree is available in Supplementary Figure S1. This phylogeny largely agreed with previous whole-genome Harveyi clade phylogenies [Thompson et al., 2009; Lin et al., 2010; Urbanczyk et al., 2013, 2014, 2016; Ke et al., 2017]. However, unlike prior studies, this phylogeny specifically aimed to resolve non-core Harveyi subclades, such as V. alginolyticus, V. antiquarius, V. diabolicus, and V. parahaemolyticus. The discovery of a well-supported V. diabolicus subclade, encompassing V. diabolicus, V. antiquarius, and certain V. alginolyticus strains, represented a novel finding.
FIGURE 1. Phylogenetic tree of the Vibrio harveyi clade. A maximum-likelihood tree representing 49 V. harveyi clade genomes was inferred from the concatenated alignment of 1,109 homologous genes. Collapsed subclades represent the core V. harveyi clade species. The V. diabolicus subclade was highlighted in blue. Node labels show the bootstrap support values. Nodes with strong support (>85) were highlighted in green while nodes with less support (<85) were highlighted in red. Branch lengths represent the average number of substitutions per site. The tree was rooted to the outgroup comprised of V. azureus NBRC 104587 and V. sagamiensis NBRC 104589.
Phylogenetic Network
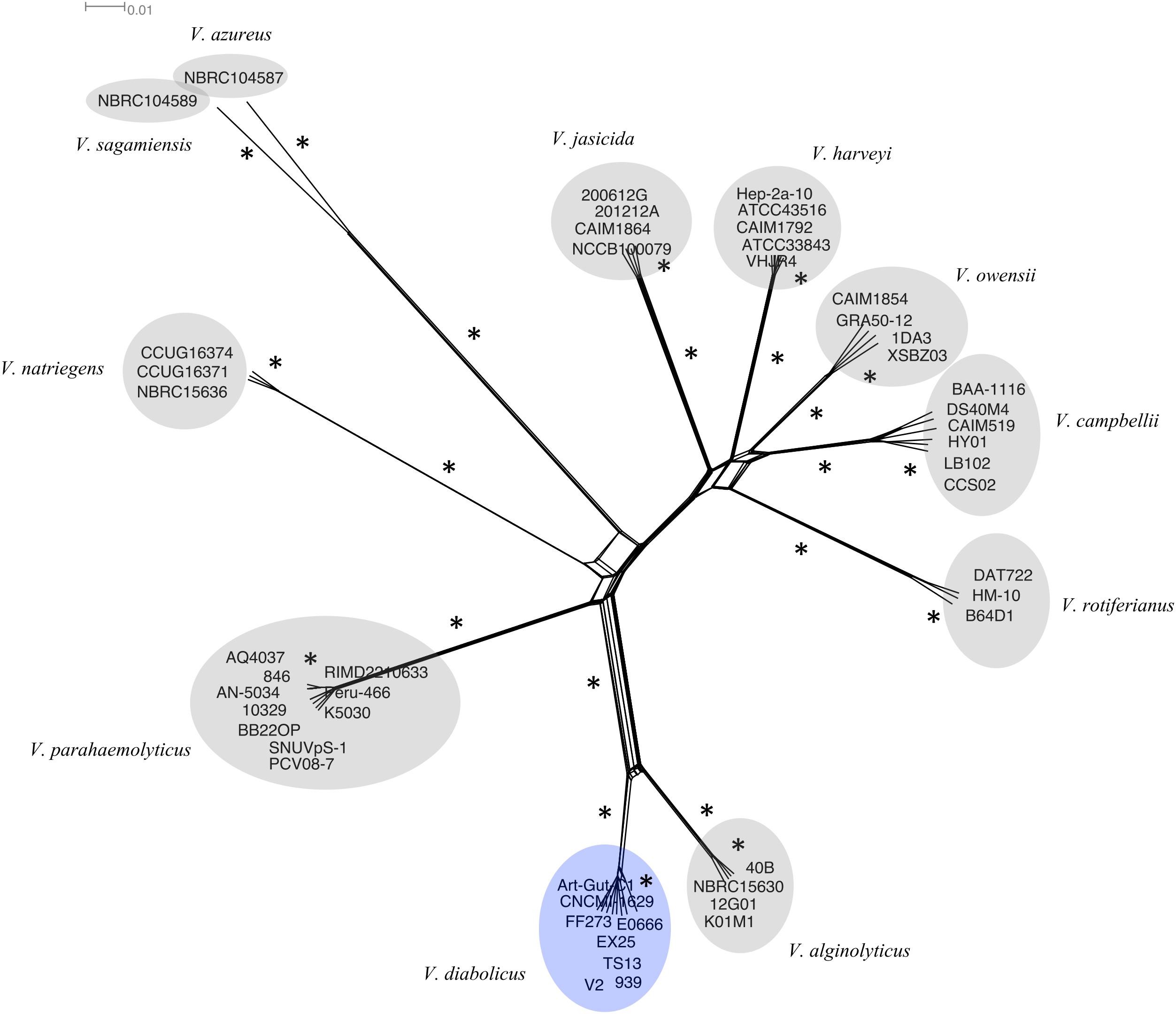
A Neighbor-Net network, constructed using the same 1,109 multilocus alignment, showed strong congruence with the phylogenetic tree (Figure 2). The network indicated a slightly more reticulate relationship between the V. diabolicus and V. alginolyticus subclades. Nevertheless, bootstrap analysis of the phylogenetic network provided robust support for the distinct V. diabolicus subclade, consistent with the phylogenetic tree results.
FIGURE 2. Neighbor-Net phylogenetic network of the V. harveyi clade. A Neighbor-net phylogenetic network representing 49 V. harveyi clade genomes was inferred from the concatenated alignment of 1,109 homologous genes. The V. diabolicus subclade was highlighted in blue while other species-specific subclades were highlighted in gray. Branches with strong bootstrap support (>85) marked with an asterisk (∗). The scale bar represents the number of substitutions per site.
Genome Similarity
Pairwise ANI values further elucidated the relatedness of the V. diabolicus subclade genomes and their closest phylogenetic neighbor, V. alginolyticus (Table 1). The eight genomes assigned to the V. diabolicus subclade exhibited high ANI values, exceeding 97%. In contrast, ANI values between members of this subclade and representative V. alginolyticus genomes (V. alginolyticus 12G01, 40B, K0M1, and NBRC 15630) were below 92%.
TABLE 1. The pairwise comparison of average nucleotide identity (ANI) values between V. diabolicus, V. antiquarius, and V. alginolyticus subclade genomes.
| V. diabolicus Art-Gut C1 | V. diabolicus CNCM I-1629 | V. antiquarius 939 | V. antiquarius EX25 | V. alginolyticus E0666 | V. alginolyticus FF273 | V. alginolyticus TS13 | V. alginolyticus V2 | V. alginolyticus 12G01 | V. alginolyticus 40B | V. alginolyticus K01M1 | V. alginolyticus NBRC 15630 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V. diabolicus Art-Gut C1 | – | 99.14 | 98.18 | 98.19 | 97.83 | 97.91 | 97.85 | 97.88 | 91.83 | 91.86 | 91.84 | 91.87 |
| V. diabolicus CNCM I-1629 | 99.14 | – | 98.15 | 98.17 | 97.81 | 97.88 | 97.83 | 97.86 | 91.81 | 91.84 | 91.82 | 91.85 |
| V. antiquarius 939 | 98.18 | 98.15 | – | 99.85 | 97.13 | 97.22 | 97.15 | 97.18 | 91.44 | 91.46 | 91.44 | 91.48 |
| V. antiquarius EX25 | 98.19 | 98.17 | 99.85 | – | 97.15 | 97.24 | 97.17 | 97.20 | 91.46 | 91.48 | 91.46 | 91.50 |
| V. alginolyticus E0666 | 97.83 | 97.81 | 97.13 | 97.15 | – | 99.69 | 99.74 | 99.71 | 91.41 | 91.43 | 91.41 | 91.45 |
| V. alginolyticus FF273 | 97.91 | 97.88 | 97.22 | 97.24 | 99.69 | – | 99.72 | 99.68 | 91.49 | 91.51 | 91.49 | 91.53 |
| V. alginolyticus TS13 | 97.85 | 97.83 | 97.15 | 97.17 | 99.74 | 99.72 | – | 99.70 | 91.43 | 91.45 | 91.43 | 91.47 |
| V. alginolyticus V2 | 97.88 | 97.86 | 97.18 | 97.20 | 99.71 | 99.68 | 99.70 | – | 91.46 | 91.48 | 91.46 | 91.50 |
| V. alginolyticus 12G01 | 91.83 | 91.81 | 91.44 | 91.46 | 91.41 | 91.49 | 91.43 | 91.46 | – | 99.81 | 99.79 | 99.80 |
| V. alginolyticus 40B | 91.86 | 91.84 | 91.46 | 91.48 | 91.43 | 91.51 | 91.45 | 91.48 | 99.81 | – | 99.78 | 99.79 |
| V. alginolyticus K01M1 | 91.84 | 91.82 | 91.44 | 91.46 | 91.41 | 91.49 | 91.43 | 91.46 | 99.79 | 99.78 | – | 99.79 |
| V. alginolyticus NBRC 15630 | 91.87 | 91.85 | 91.48 | 91.50 | 91.45 | 91.53 | 91.47 | 91.50 | 99.80 | 99.79 | 99.79 | – |
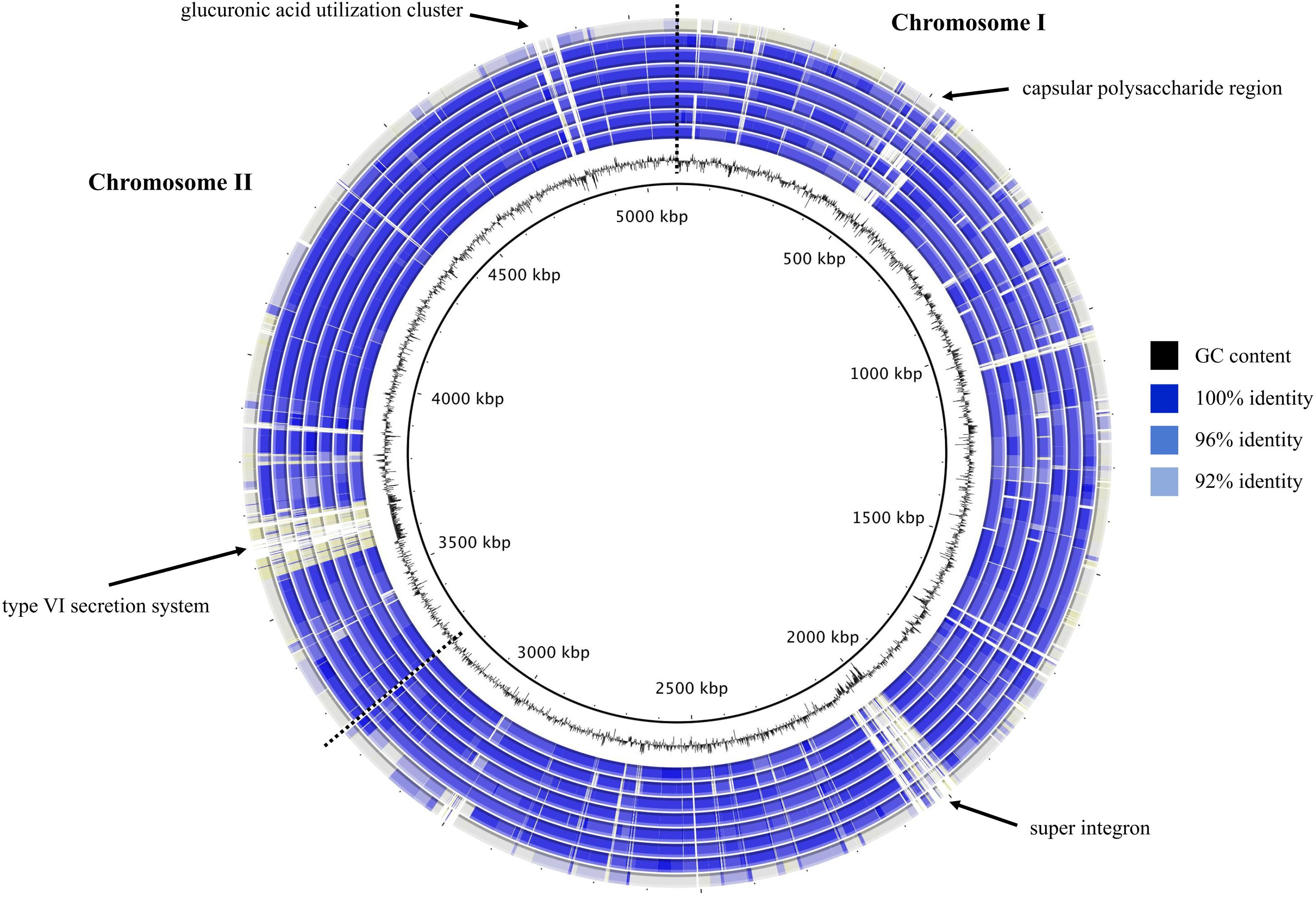
To visualize the high degree of synonymy indicated by ANI values, a circular blastn map was constructed (Figure 3). This map clearly illustrated the extensive sequence similarity among the two V. diabolicus genomes, the two V. antiquarius genomes, and the four V. alginolyticus genomes, all relative to the V. antiquarius EX25 reference genome. In contrast, the map highlighted the sequence divergence between these genomes and V. alginolyticus NBRC 15630. Notable hypervariable genomic regions identified on the map included a capsular polysaccharide region, a super integron, a type VI secretion system (T6SS), and a glucuronic acid utilization cluster.
FIGURE 3. Blastmap comparison of V. diabolicus subclade genomes. A circular comparison of the eight V. diabolicus subclade genomes was based on the blastn analysis of whole genomes. Starting at the center, concentric rings show the sequence similarity between the query genome (V. diabolicus Art-Gut C1, V. diabolicus CNCM I-1629, V. antiquarius 939, V. alginolyticus TS13, V. alginolyticus FF273, V. alginolyticus V2, V. alginolyticus E0666, and V. alginolyticus NBRC 15630, respectively) and the reference genome (V. antiquarius EX25). Blue shading depicts regions with 100, 96, and 92% sequence similarity. Regions with less than 92% sequence similarity shown in gray. Dashed lines partition chromosomes I and II.
A broader comparison of the eight V. diabolicus subclade isolates (Supplementary Table S1) revealed their diverse origins, spanning various isolation sources and geographic locations. Genome sizes ranged from 5,048,917 bp (V. alginolyticus E0666) to 5,430,661 bp (V. antiquarius 939). Gene counts varied from 4,572 (V. alginolyticus FF273) to 5,251 (V. diabolicus CNCM I-1629), and GC content ranged from 44.6% (V. alginolyticus FF273) to 44.9% (V. antiquarius EX25) (Table 2).
TABLE 2. A summary of the attributes of the V. diabolicus subclade genomes: V. diabolicus Art-Gut C1 and CNCM I-1629, V. antiquarius 939 and EX25, and V. alginolyticus E0666, FF273, TS13 and V2.
| Genome | Genome Size (bp) | %GC Content | Total Genes | Protein Coding Genes | RNA Genes | Pseudogenes |
|---|---|---|---|---|---|---|
| V. diabolicus Art-Gut C1 | 5,341,424 | 44.8 | 5,048 | 4,876 | 111 | 61 |
| V. diabolicus CNCM I-1629 | 5,354,801 | 44.8 | 5,251 | 5,075 | 115 | 61 |
| V. antiquarius 939 | 5,430,661 | 44.7 | 5,049 | 4,876 | 112 | 61 |
| V. antiquarius EX25 | 5,340,885 | 44.9 | 5,038 | 4,865 | 112 | 61 |
| V. alginolyticus E0666 | 5,048,917 | 44.8 | 4,684 | 4,521 | 102 | 61 |
| V. alginolyticus FF273 | 5,148,508 | 44.6 | 4,572 | 4,413 | 99 | 60 |
| V. alginolyticus TS13 | 5,240,985 | 44.7 | 4,785 | 4,619 | 105 | 61 |
| V. alginolyticus V2 | 5,180,348 | 44.7 | 4,778 | 4,617 | 100 | 61 |
Genome Diversity
Pangenome analysis, using both get_homologues and Panseq, was conducted to assess genome diversity within the V. diabolicus subclade. Get_homologues analysis identified a total pangenome of 4,854 genes, with 3,422 core genes shared by all eight genomes (approximately 70.5% of the pangenome) and 1,422 accessory genes (approximately 29.5%). Panseq analysis estimated a larger pangenome size of 7,856,358 bp, with a 4,605,287 bp core genome (approximately 58.6%) and a 3,251,071 bp accessory genome (approximately 41.4%).
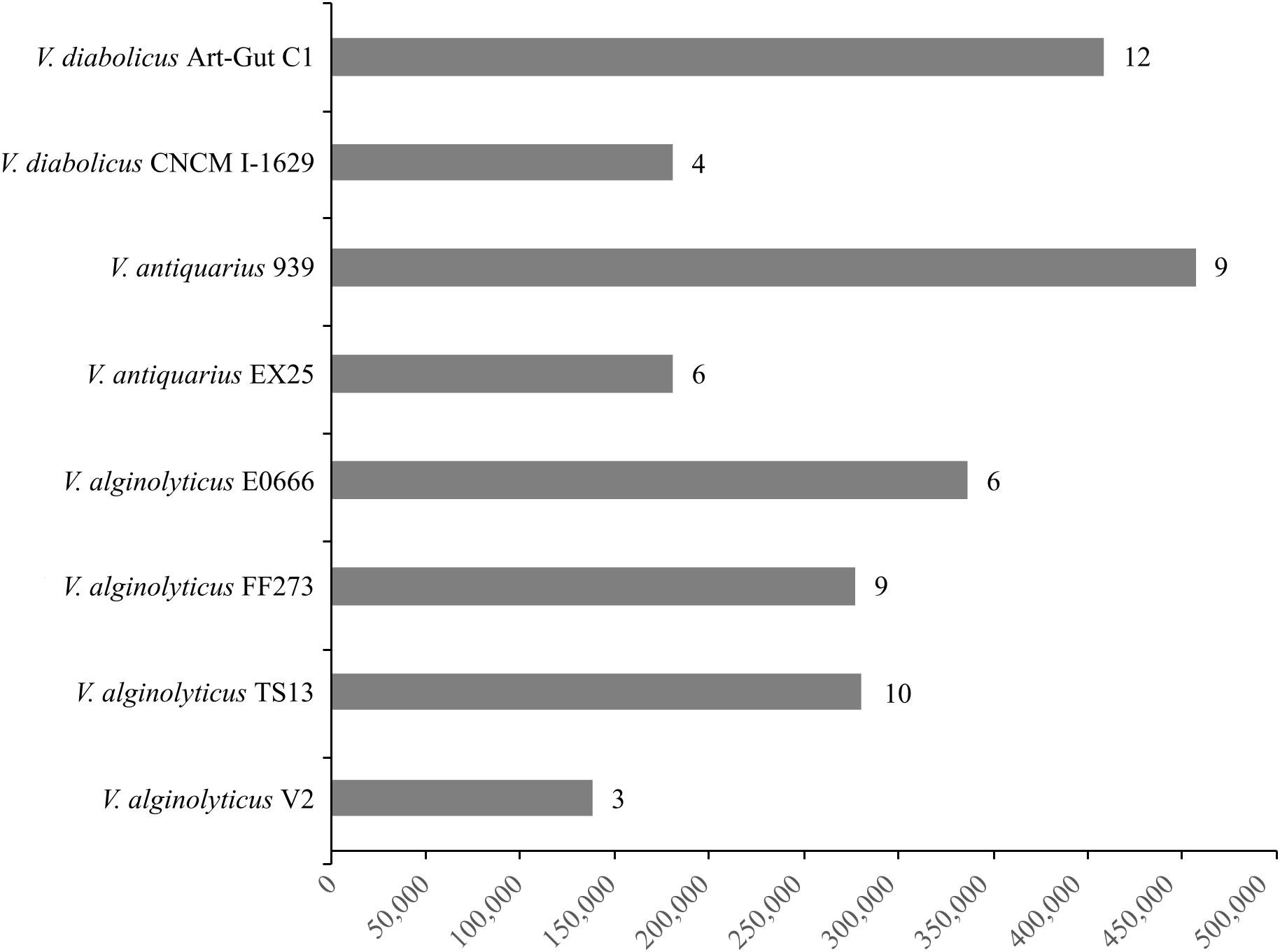
Panseq’s Novel Region Finder revealed substantial novel genomic regions, totaling 2,295,535 bp across all eight genomes, accounting for the majority of the accessory genome. Probable genomic islands (GIs) larger than 10 Kb were prevalent within these novel regions (Figure 4). These GIs encoded a variety of proteins, including hypothetical proteins, capsular and lipopolysaccharide biosynthesis proteins, carbohydrate and DNA metabolism proteins, membrane transport proteins, phage-related proteins, and conjugative transfer proteins. However, these novel regions and GIs did not encode proteins with obvious habitat-specific functions but rather features associated with broader environmental fitness. Notably, a V. alginolyticus TS13 GI (26,181 bp) carried a widespread colonization island (WCI) with a tight-adherence (tad) locus. The alkyl hydroperoxide reductase, previously suggested to be involved in deep-sea hydrogen peroxide scavenging, was found in all eight V. diabolicus strains.
FIGURE 4. Novel regions and genomic islands (GIs). The cumulative length (bp) of novel genomic regions present in each V. diabolicus subclade genome. The bars are labeled with the number of probable GIs (i.e., novel genomic regions larger than 10 Kb) present in each genome.
Subclade-Associated Genes
Screening for genes ubiquitous in the V. diabolicus subclade but absent from the other 41 strains identified five genes (Table 3). These genes were single-copy, highly conserved, and exhibited greater than 96% sequence identity in both gene and protein alignments. Two genes encoded hypothetical proteins (chromosome I), while the remaining three encoded a hydroxyectoine utilization dehydratase, an ornithine cyclodeaminase, and a phosphoesterase (chromosome II).
TABLE 3. Description of open reading frames (ORFs) ubiquitous in V. diabolicus subclade strains (i.e., detected in V. diabolicus Art-Gut C1 and CNCM I-1629, V. antiquarius 939 and EX25, and V. alginolyticus E0666, FF273, TS13, and V2) but absent from the 41 other strains included in this study.
| Locus Tag | Gene Product | Chromosome | Average Sequence Identity (%) |
|---|---|---|---|
| VANT939_01458 | Hypothetical protein | I | 97.8 |
| VANT939_01459 | Hypothetical protein | I | 96.4 |
| VANT939_02888 | Hydroxyectoine utilization dehydratase | II | 98.8 |
| VANT939_02919 | Ornithine cyclodeaminase | II | 98.6 |
| VANT939_03583 | Phosphoesterase | II | 98.9 |
Metagenome Survey
Metagenome database queries using three subclade-associated coding sequences (hydroxyectoine utilization dehydratase, ornithine cyclodeaminase, and phosphoesterase) as markers detected homologous proteins in over 100 environmental metagenomes, predominantly marine. However, the sequence identity of the highest-scoring hits was not comparable to the subclade protein alignments (less than 95%), indicating non-significant matches.
Potential Virulence
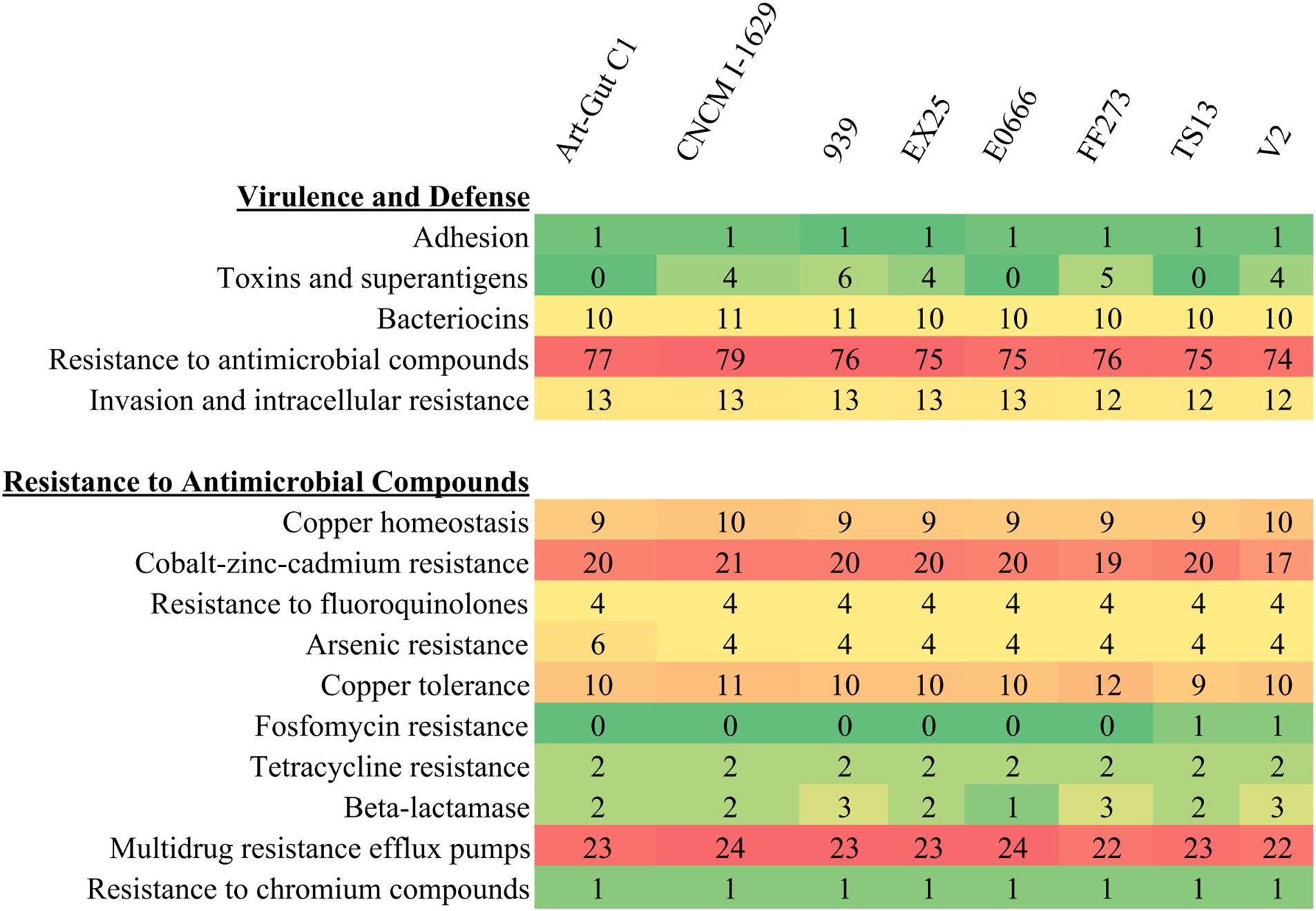
SEED subsystem feature comparisons revealed a substantial repertoire of virulence and defense-related features across the V. diabolicus subclade genomes (Figure 5). Subsystem profiles were remarkably consistent among all eight genomes. For instance, the accessory colonization factor AcfA (Adhesion subsystem) and a colicin V production cluster (Bacteriocins and Ribosomally Synthesized Antibacterial Peptides subsystem, 10–11 features) were present in all genomes. Similarly, a suite of proteins homologous to a Mycobacterium virulence operon involved in protein synthesis (Invasion and Intracellular Resistance subsystem, 12–13 features) was also conserved. The Toxins and Superantigens subsystem showed some variability (0–6 features), but when present, included proteins like the transcriptional activator ToxR, transmembrane regulatory protein ToxS, Zonula occludens toxin (Zot), and accessory cholera enterotoxin (Ace).
FIGURE 5. Heatmap of SEED subsystems. The number of SEED subsystem feature counts related to Virulence and Defense (top) with focus on subsystem feature counts related to Resistance to Antimicrobial Compounds (bottom). Coloring correlates with the prevalence of specific features with rare features appearing green and abundant features appearing red.
Analysis of the Resistance to Antimicrobial Compounds subsystem revealed 74–79 features per genome, with a nearly identical resistome across all eight genomes. This resistome included features associated with resistance to toxic metals and metalloids (copper, cobalt, zinc, cadmium, chromium, arsenic) and antibiotics (fluoroquinolones, fosfomycin, tetracycline, penicillin). Additionally, 22–24 efflux pumps per genome were identified, belonging to the multidrug and toxin extrusion (MATE) and resistance-nodulation-cell division (RND) families.
Phylogenetic analysis of β-lactamase proteins (1–3 per genome) indicated two distinct β-lactamase classes (Supplementary Figure S2): class A and class C, associated with resistance to carbenicillin and cephalosporin, respectively, based on ARDB annotations. FosA enzymes, conferring fosfomycin resistance and present in V. alginolyticus TS13 and V2, were identified as identical glutathione transferases.
Antibiotic Susceptibility
The Cefinase test confirmed β-lactamase production in both V. antiquarius 939 and V. parahaemolyticus RIMD2210633A. V. antiquarius 939 exhibited resistance to penicillin, ampicillin, and cephalothin, but susceptibility to carbenicillin. V. parahaemolyticus RIMD2210633 showed resistance to penicillin and cephalothin, intermediate susceptibility to ampicillin, and susceptibility to carbenicillin.
Discussion
The taxonomy of the Vibrio genus is characterized by rapid expansion and constant revision. The number of recognized species has dramatically increased in recent decades, from five [Bergey et al., 1974] to the current count of 139. Species boundaries within the Harveyi clade, encompassing V. campbellii, V. harveyi, V. rotiferianus, V. natriegens, and V. parahaemolyticus, have been particularly challenging to define [Thompson et al., 2006]. However, advances in multilocus [Thompson et al., 2005, 2007; Sawabe et al., 2007, 2013; González-Escalona et al., 2008] and genome-scale phylogenetics [Thompson et al., 2009; Lin et al., 2010; Urbanczyk et al., 2013, 2014, 2016; Ke et al., 2017] have significantly improved our understanding of this complex clade. This study builds upon this foundation by clarifying the taxonomic relationships of the deep-sea species V. diabolicus and V. antiquarius.
The initial impetus for this research was to resolve the taxonomic identity of Vibrio sp. 939. Genome sequencing and subsequent genome-scale taxonomic analysis of 49 Harveyi clade genomes were undertaken. Initial RAST analysis suggested a close relationship to V. antiquarius, leading to the V. antiquarius 939 GenBank designation. However, a more detailed maximum-likelihood 49-genome phylogeny, based on 1,109 homologous genes, placed the isolate within a monophyletic subclade. This subclade unexpectedly included not only V. diabolicus and V. antiquarius strains but also four strains initially identified as V. alginolyticus.
Previous studies have highlighted the challenges in defining species boundaries within the Harveyi clade due to complex evolutionary processes like recombination, which can obscure phylogenetic signals [Fraser et al., 2007; Thompson et al., 2007]. To address this, a phylogenetic network was constructed to visualize the uncertainty in species boundaries. The network revealed short branching between V. jasicida, V. harveyi, V. owensii, and V. campbellii, and between V. alginolyticus and V. diabolicus, suggesting close relationships and potential for genetic exchange. A pronounced reticulation pattern between V. alginolyticus and V. diabolicus further indicated that recombination might be blurring the phylogenetic distinction between these two species. Despite these complexities, the subclades identified by the network analysis were well-supported and consistent with the maximum-likelihood phylogeny.
The 95–96% ANI threshold is widely used as a benchmark for prokaryotic species delineation [Richter and Rosselló-Móra, 2009; Varghese et al., 2015]. In this study, the observation that V. diabolicus, V. antiquarius, and the four V. alginolyticus genomes shared greater than 97% ANI strongly suggests that they belong to the same species. Conversely, their ANI values below 92% when compared to representative V. alginolyticus genomes confirmed their distinctness from this closely related species. Analogous to a previous study that used a 97% ANI threshold to establish V. inhibens as a heterotypic synonym of V. jasicida [Urbanczyk et al., 2016], our ANI analysis clearly demonstrates that V. antiquarius and the four V. alginolyticus strains are synonyms of V. diabolicus. This finding highlights the importance of comparative genomics in resolving taxonomic ambiguities and identifying synonyms in bacterial classification.
The diverse origins of the eight V. diabolicus strains, isolated from various sources and geographic locations, imply a broad range of physiological capabilities. Genomic variability is considered a key factor enabling prokaryotic species to thrive in diverse habitats [Cordero and Polz, 2014]. The substantial genomic variability observed within V. diabolicus, reflected in its large accessory genome and numerous strain-specific GIs, may correlate with its ability to persist in such varied environments globally. For example, the tad locus-encoded Flp pilus, found on a GI in V. alginolyticus TS13, could facilitate colonization of diverse substrates and biofilm formation, crucial for bacterial persistence [Kachlany et al., 2001]. However, the species appears to be “conditionally rare,” as species-specific genetic markers developed in this study were not detected in environmental metagenome databases.
The taxonomic synonymy of V. antiquarius with V. diabolicus represents a significant shift in the understanding of these bacteria, previously considered distinct species [Raguénès et al., 1997; Goudenège et al., 2014; Hasan et al., 2015]. The most striking synonym, however, is V. alginolyticus E0666, isolated from spoiled horse mackerel and shown to be virulent in a murine model [Cao et al., 2013; Liu et al., 2013]. The presence of virulence factors like Ace and Zot in V. antiquarius EX25 was previously reported [Hasan et al., 2015]. This study confirmed the widespread presence of these and other virulence factors across the expanded V. diabolicus subclade, suggesting that virulence-associated genes may be ecologically relevant beyond pathogenicity, potentially playing a broader role in environmental fitness, as hypothesized previously [Hasan et al., 2015].
The intrinsic antibiotic resistance of V. diabolicus strains, potentially complicating treatment in case of virulence, is also noteworthy. Resistome analysis revealed genes associated with resistance to fluoroquinolones, fosfomycin, tetracycline, and penicillin. Experimentally confirmed β-lactamase production and resistance to penicillin, ampicillin, and cephalosporin in V. antiquarius 939 further underscore this concern. The abundance of efflux pumps, including MATE and RND families, could expand resistance mechanisms. The ubiquity of these resistance features, even in a species considered rare and previously associated with isolated deep-sea environments, is concerning, especially given the increasing antibiotic resistance trends in other Vibrio species like V. cholerae, V. parahaemolyticus, and V. vulnificus [Garg et al., 2000; Baker-Austin et al., 2008, 2009; Devi et al., 2009; Shaw et al., 2014; Uppal et al., 2017].
Conclusion
This taxonomic and comparative genomic analysis conclusively demonstrates that V. diabolicus and V. antiquarius should be reclassified as a single species. Given the earlier discovery and publication of V. diabolicus [Raguénès et al., 1997; Goudenège et al., 2014] compared to V. antiquarius [Hasan et al., 2015], V. antiquarius is formally designated as a taxonomic synonym of V. diabolicus. V. alginolyticus remains a distinct species, but the four misidentified strains (E0666, FF273, TS13, and V2) are now recognized as synonyms of *V. diabolicus. The expansion of the V. diabolicus species is ongoing, with the recent public release of three additional V. diabolicus genomes (FDAARGOS_96, LMG 3418, and FDAARGOS_105) during the review of this manuscript, further solidifying this revised taxonomic understanding. This study underscores the dynamic nature of bacterial taxonomy and the power of comparative** genomics in refining our understanding of microbial diversity and species boundaries.
Author Contributions
JWT, WN, RP, EA, and MS conceived and designed the research project. JWT, JJT, AM, LP, NE, and DNA conducted the genomic analyses and laboratory experiments. All authors contributed to data interpretation and the writing of the manuscript.
Funding
This study was funded by the National Oceanic and Atmospheric Administration’s (NOAA) Oceans and Human Health Initiative (OHHI), the National Marine Fisheries Service (NMFS), the National Research Council (NRC), the Gordon and Betty Moore Foundation (EVA #537.01), and Texas A&M University-Corpus Christi (TAMU-CC).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study would not have been possible without the support of our friends and colleagues. In particular, we thank Chris T. Berthiaume, Rhonda Morales, and Thomas Merrick for their assistance with sequencing and data analysis, and we thank Drs. Toshio Kodama and Tetsuya Iida and the Pathogenic Microbes Repository unit of the Research Institute for Microbial Diseases at Osaka University for providing the V. parahaemolyticus RIMD2210633 strain. We also thank Dr. Daniele Provenzano for kindly proofreading the manuscript and Dr. Magesh Thiyagarajan for his generous gift of the laboratory supplies used during this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01893/full#supplementary-material
FIGURE S1 | Phylogenetic tree of the V. harveyi clade. A maximum-likelihood tree representing 49 V. harveyi clade genomes was inferred from the concatenated alignment of 1,109 homologous genes. Node labels show the bootstrap support values. Nodes with strong support (>85) were highlighted in green while nodes with weak support (<85) were highlighted in red. Branch lengths represent the average number of substitutions per site. The tree was rooted to the outgroup comprised of V. azureus NBRC104587 and V. sagamiensis NBRC 104589.
FIGURE S2 | Phylogenetic tree of β-lactamases. A maximum-likelihood tree showing the relatedness of 18 β-lactamases present in the eight V. diabolicus subclade genomes. Node labels show the bootstrap support values greater than 85. Branch lengths represent the average number of substitutions per site. The tree was rooted at the midpoint.
TABLE S1 | List of the 49 Harveyi clade genomes included in this study. Genomes in bold font belong to the new V. diabolicus subclade described in this study.
Footnotes
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
PubMed Abstract | CrossRef Full Text | Google Scholar
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Austin, B., and Zhang, X. H. (2006). Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43, 119–124. doi: 10.1111/j.1472-765X.2006.01989.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75–95. doi: 10.1186/1471-2164-9-75
PubMed Abstract | CrossRef Full Text | Google Scholar
Baker-Austin, C., McArthur, J. V., Lindell, A. H., Wright, M. S., Tuckfield, R. C., Gooch, J., et al. (2009). Multisite analysis reveals widespread antibiotic resistance in the marine pathogen Vibrio vulnificus. Microb. Ecol. 57, 151–159. doi: 10.1007/s00248-008-9413-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Baker-Austin, C., McArthur, J. V., Tuckfield, R. C., Najarro, M., Lindell, A. H., Gooch, J., et al. (2008). Antibiotic resistance in the shellfish pathogen Vibrio parahaemolyticus isolated from the coastal water and sediment of Georgia and South Carolina, USA. J. Food Prot. 71, 2552–2558. doi: 10.4315/0362-028X-71.12.2552
PubMed Abstract | CrossRef Full Text | Google Scholar
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Bergey, D. H., Buchanan, R. E., and Gibbons, N. E. (1974). Bergey’s manual of determinative bacteriology, 8th Edn, vol. 1. Baltimore, MD: Williams & Wilkins Co.
Bryant, D., and Moulton, V. (2004). Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol. Biol. Evol. 21, 255–265. doi: 10.1093/molbev/msh018
PubMed Abstract | CrossRef Full Text | Google Scholar
Cao, Y., Liu, X. F., Zhang, H. L., Chen, Y. J., and Hu, C. J. (2013). Draft genome sequence of the human-pathogenic bacterium Vibrio alginolyticus E0666. Genome Announc. 1:e686-13. doi: 10.1128/genomeA.00686-13
PubMed Abstract | CrossRef Full Text | Google Scholar
Capella-Gutierrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
PubMed Abstract | CrossRef Full Text | Google Scholar
Chakraborty, S., Nair, G. B., and Shinoda, S. (1997). Pathogenic Vibrios in the natural aquatic environment. Rev. Environ. Health 12, 63–80. doi: 10.1515/REVEH.1997.12.2.63
CrossRef Full Text | Google Scholar
Chikhi, R., and Medvedev, P. (2014). Informed and automated k-mer size selection for genome assembly. Bioinformatics 30, 31–37. doi: 10.1093/bioinformatics/btt310
PubMed Abstract | CrossRef Full Text | Google Scholar
Clinical and Laboratory Standards Institute [CLSI] (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria, Approved Guideline, 2nd Edn. Wayne, IL: Clinical Laboratory Standards Institute.
Contreras-Moreira, B., and Vinuesa, P. (2013). GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 79, 7696–7701. doi: 10.1128/AEM.02411-13
PubMed Abstract | CrossRef Full Text | Google Scholar
Cordero, O. X., and Polz, M. F. (2014). Explaining microbial genomic diversity in light of evolutionary ecology. Nat. Rev. Microbiol. 12, 263–273. doi: 10.1038/nrmicro3218
PubMed Abstract | CrossRef Full Text | Google Scholar
Devi, R., Surendran, P. K., and Chakraborty, K. (2009). Antibiotic resistance and plasmid profiling of Vibrio parahaemolyticus isolated from shrimp farms along the southwest coast of India. World J. Microbiol. Biotechnol. 25, 2005–2012. doi: 10.1007/s11274-009-0101-8
CrossRef Full Text | Google Scholar
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
PubMed Abstract | CrossRef Full Text | Google Scholar
Frans, I., Michiels, C. W., Bossier, P., Willems, K. A., Lievens, B., and Rediers, H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34, 643–661. doi: 10.1111/j.1365-2761.2011.01279.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Fraser, C., Hanage, W. P., and Spratt, B. G. (2007). Recombination and the nature of bacterial speciation. Science 315, 476–480. doi: 10.1126/science.1127573
PubMed Abstract | CrossRef Full Text | Google Scholar
Garg, P., Chakraborty, S., Basu, I., Datta, S., Rajendran, K., Bhattacharya, T., et al. (2000). Expanding multiple antibiotic resistance among clinical strains of Vibrio cholerae isolated from 1992–1997 in Calcutta, India. Epidemiol. Infect. 124, 393–399. doi: 10.1017/S0950268899003957
PubMed Abstract | CrossRef Full Text | Google Scholar
Giardine, B., Riemer, C., Hardison, R. C., Burhans, R., Elnitski, L., Shah, P., et al. (2005). Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455. doi: 10.1101/gr.4086505
PubMed Abstract | CrossRef Full Text | Google Scholar
González-Escalona, N., Martinez-Urtaza, J., Romero, J., Espejo, R. T., Jaykus, L. A., and DePaola, A. (2008). Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190, 2831–2840. doi: 10.1128/JB.01808-07
PubMed Abstract | CrossRef Full Text | Google Scholar
Goudenège, D., Boursicot, V., Versigny, T., Bonnetot, S., Ratiskol, J., Sinquin, C., et al. (2014). Genome sequence of Vibrio diabolicus and identification of the exopolysaccharide HE800 biosynthesis locus. Appl. Microbiol. Biotechnol. 98, 10165–10176. doi: 10.1007/s00253-014-6086-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Hasan, N. A., Grim, C. J., Lipp, E. K., Rivera, I. N., Chun, J., Haley, B. J., et al. (2015). Deep-sea hydrothermal vent bacteria related to human pathogenic Vibrio species. Proc. Natl. Acad. Sci. U.S.A. 112, E2813–E2819. doi: 10.1073/pnas.1503928112
PubMed Abstract | CrossRef Full Text | Google Scholar
Huson, D. H., and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
PubMed Abstract | CrossRef Full Text | Google Scholar
Johnson, C. N., Bowers, J. C., Griffitt, K. J., Molina, V., Clostio, R. W., Pei, S., et al. (2012). Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microbiol. 78, 7249–7257. doi: 10.1128/AEM.01296-12
PubMed Abstract | CrossRef Full Text | Google Scholar
Kachlany, S. C., Planet, P. J., DeSalle, R., Fine, D. H., and Figurski, D. H. (2001). Genes for tight adherence of Actinobacillus actinomycetemcomitans: from plaque to plague to pond scum. Trends Microbiol. 9, 429–437. doi: 10.1016/S0966-842X(01)02161-8
PubMed Abstract | CrossRef Full Text | Google Scholar
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
PubMed Abstract | CrossRef Full Text | Google Scholar
Karaolis, D. K., Lan, R., and Reeves, P. R. (1995). The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177, 3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995
PubMed Abstract | CrossRef Full Text | Google Scholar
Ke, H. M., Prachumwat, A., Yu, C. P., Yang, Y. T., Promsri, S., Liu, K. F., et al. (2017). Comparative genomics of Vibrio campbellii strains and core species of the Vibrio harveyi clade. Sci. Rep. 7, 41394–41311. doi: 10.1038/srep41394
PubMed Abstract | CrossRef Full Text | Google Scholar
Klein, S. L., Gutierrez-West, C. K., Mejia, D. M., and Lovell, C. R. (2014). Genes similar to the Vibrio parahaemolyticus virulence-related genes tdh, tlh, and vscC2 occur in other Vibrionaceae species isolated from a pristine estuary. Appl. Environ. Microbiol. 80, 595–602. doi: 10.1128/AEM.02895-13
PubMed Abstract | CrossRef Full Text | Google Scholar
Klimke, W., Agarwala, R., Badretdin, A., Chetvernin, S., Ciufo, S., Fedorov, B., et al. (2009). The national center for biotechnology information’s protein clusters database. Nucleic Acids Res. 37, D216–D223. doi: 10.1093/nar/gkn734
PubMed Abstract | CrossRef Full Text | Google Scholar
Laing, C., Buchanan, C., Taboada, E. N., Zhang, Y., Kropinski, A., Villegas, A., et al. (2010). Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics 11:461. doi: 10.1186/1471-2105-11-461
PubMed Abstract | CrossRef Full Text | Google Scholar
Letchumanan, V., Pusparajah, P., Tan, L. T.-H., Yin, W.-F., Lee, L.-H., and Chan, K.-G. (2015). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Micriobiol. 6:1417. doi: 10.3389/fmicb.2015.01417
PubMed Abstract | CrossRef Full Text | Google Scholar
Li, L., Stoeckert, C. J., and Roos, D. S. (2003). OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189. doi: 10.1101/gr.1224503
PubMed Abstract | CrossRef Full Text | Google Scholar
Lin, B., Wang, Z., Malanoski, A. P., O’Grady, E. A., Wimpee, C. F., Vuddhakul, V., et al. (2010). Comparative genomic analyses identify the Vibrio harveyi genome sequenced strains BAA-1116 and HY01 as Vibrio campbellii. Environ. Microbiol. Rep. 2, 81–89. doi: 10.1111/j.1758-2229.2009.00100.x
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, B., and Pop, M. (2009). ARDB-antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
PubMed Abstract | CrossRef Full Text | Google Scholar
Liu, X. F., Zhang, H., Liu, X., Gong, Y., Chen, Y., Cao, Y., et al. (2013). Pathogenic analysis of Vibrio alginolyticus infection in a mouse model. Folia Microbiol. 59, 167–171. doi: 10.1007/s12223-013-0279-x
PubMed Abstract | CrossRef Full Text | Google Scholar
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
PubMed Abstract | CrossRef Full Text | Google Scholar
Markert, S., Arndt, C., Felbeck, H., Becher, D., Sievert, S. M., Hügler, M., et al. (2007). Physiological proteomics of the uncultured endosymbiont of Riftia pachyptila. Science 315, 247–250. doi: 10.1126/science.1132913
PubMed Abstract | CrossRef Full Text | Google Scholar
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
CrossRef Full Text | Google Scholar
Matsumoto, C., Okuda, J., Ishibashi, M., Iwanaga, M., Garg, P., Rammamurthy, T., et al. (2000). Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38, 578–585.
PubMed Abstract | Google Scholar
Minh, B. Q., Nguyen, M. A. T., Haeseler, and von, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
PubMed Abstract | CrossRef Full Text | Google Scholar
Nasu, H., Iida, T., Sugahara, T., Yamaichi, Y., Park, K.-S., Yokoyama, T., et al. (2000). A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38, 2156–2161.
PubMed Abstract | Google Scholar
Nguyen, L. T., Schmidt, H. A., Haeseler, von, A., and Minh, B. Q. (2014). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
PubMed Abstract | CrossRef Full Text | Google Scholar
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
PubMed Abstract | CrossRef Full Text | Google Scholar
Pacini, F. (1854). Osservazioni microscopiche e deduzioni patologiche sul cholera asiatico. Gaz. Med. Ital. Toscana 6, 397–405.
Paranjpye, R., Hamel, O. S., Stojanovski, A., and Liermann, M. (2012). Genetic diversity of clinical and environmental Vibrio parahaemolyticus strains from the Pacific Northwest. Appl. Environ. Microbiol. 78, 8631–8638. doi: 10.1128/AEM.01531-12
PubMed Abstract | CrossRef Full Text | Google Scholar
Polz, M. F., Hunt, D. E., Preheim, S. P., and Weinreich, D. M. (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 2009–2021. doi: 10.1098/rstb.2006.1928
PubMed Abstract | CrossRef Full Text | Google Scholar
Raguénès, G., Christen, R., Guezennac, J., Pignet, P., and Barbier, G. (1997). Vibrio diabolicus sp. nov., a new polysaccharide-secreting organism isolated from a deep-sea hydrothermal vent polychaete annelid, Alvinella pompejana. Int. J. Syst. Bacteriol. 47, 989–995. doi: 10.1099/00207713-47-4-989
PubMed Abstract | CrossRef Full Text | Google Scholar
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
PubMed Abstract | CrossRef Full Text | Google Scholar
Richter, M., Rosselló-Móra, R., Oliver Glöckner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
PubMed Abstract | CrossRef Full Text | Google Scholar
Rosenberg, E., Koren, O., Reshef, L., Efrony, R., and Zilber-Rosenberg, I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5, 355–362. doi: 10.1038/nrmicro1635
PubMed Abstract | CrossRef Full Text | Google Scholar
Sawabe, T., Kita-Tsukamoto, K., and Thompson, F. L. (2007). Inferring the evolutionary history of Vibrios by means of multilocus sequence analysis. J. Bacteriol. 189, 7932–7936. doi: 10.1128/JB.00693-07
PubMed Abstract | CrossRef Full Text | Google Scholar
Sawabe, T., Ogura, Y., Matsumura, Y., Feng, G., Mino, S., Nakagawa, S., et al. (2013). Updating the Vibrio clades defined by multilocus sequence phylogeny: proposal of eight new clades, and the description of Vibrio tritonius sp. nov. Front. Microbiol. 4:414. doi: 10.3389/fmicb.2013.00414
CrossRef Full Text | Google Scholar
Shade, A., Jones, S. E., Caporaso, J. G., Handelsman, J., Knight, R., Fierer, N., et al. (2014). Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 5:e1371-14. doi: 10.1128/mBio.01371-14
PubMed Abstract | CrossRef Full Text | Google Scholar
Shaw, K. S., Rosenberg Goldstein, R. E., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of Chesapeake Bay and Maryland coastal bays. PLoS One 9:e89616-11. doi: 10.1371/journal.pone.0089616
PubMed Abstract | CrossRef Full Text | Google Scholar
Strom, M. S., and Paranjpye, R. N. (2000). Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2, 177–188. doi: 10.1016/S1286-4579(00)00270-7
PubMed Abstract | CrossRef Full Text | Google Scholar
Takemura, A. F., Chien, D. M., and Polz, M. F. (2014). Associations and dynamics of Vibrionaceae in the environment, from the genus to the population level. Front. Microbiol. 5:38. doi: 10.3389/fmicb.2014.00038
CrossRef Full Text | Google Scholar
Thompson, C. C., Vicente, A., Souza, R. C., Vasconcelos, A., Vesth, T., Alves, N., et al. (2009). Genomic taxonomy of Vibrios. BMC Evol. Biol. 9:258–273. doi: 10.1186/1471-2148-9-258
PubMed Abstract | CrossRef Full Text | Google Scholar
Thompson, F. L., Austin, B., and Swings, J. E. (2006). “Taxonomy of the vibrios,” in The Biology of Vibrios, eds F. L. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press), 29–43. doi: 10.1128/9781555815714.ch3
CrossRef Full Text | Google Scholar
Thompson, F. L., Gevers, D., Thompson, C. C., Dawyndt, P., Naser, S., Hoste, B., et al. (2005). Phylogeny and molecular identification of Vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71, 5107–5115. doi: 10.1128/AEM.71.9.5107-5115.2005
PubMed Abstract | CrossRef Full Text | Google Scholar
Thompson, F. L., Gomez-Gil, B., Vasconcelos, A. T., and Sawabe, T. (2007). Multilocus sequence analysis reveals that Vibrio harveyi and V. campbellii are distinct species. Appl. Environ. Microbiol. 73, 4279–4285. doi: 10.1128/AEM.00020-07
PubMed Abstract | CrossRef Full Text | Google Scholar
Thompson, F. L., Iida, T., and Swings, J. (2004). Biodiversity of Vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. doi: 10.1128/MMBR.68.3.403-431.2004
PubMed Abstract | CrossRef Full Text | Google Scholar
Thompson, J. R., and Polz, M. F. (2006). “Dynamics of Vibrio populations and their role in environmental nutrient cycling,” in The Biology of Vibrios, eds F. L. Thompson, B. Austin, and J. Swings (Washington, DC: ASM Press), 190–203.
Turner, J. W., Berthiaume, C. T., Morales, R., Armbrust, E. V., and Strom, M. S. (2016). Genomic evidence of adaptive evolution in emergent Vibrio parahaemolyticus ecotypes. Elem. Sci. Anth 4:117. doi: 10.12952/journal.elementa.000117
CrossRef Full Text | Google Scholar
Turner, J. W., Paranjpye, R. N., Landis, E. D., Biryukov, S. V., Gonzalez-Escalona, N., Nilsson, W. B., et al. (2013). Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8:e55726. doi: 10.1371/journal.pone.0055726
PubMed Abstract | CrossRef Full Text | Google Scholar
Uppal, B., Mehra, B., Panda, P. S., and Kumar, S. K. (2017). Changing epidemiology and antimicrobial resistance pattern of Vibrio cholerae isolates at a tertiary care health laboratory in North India (2011–2015). Trop. J. Med. Res. 20, 132–137. doi: 10.4103/tjmr.tjmr_17_17
CrossRef Full Text | Google Scholar
Urbanczyk, H., Ogura, Y., and Hayashi, T. (2013). Taxonomic revision of Harveyi clade bacteria (family Vibrionaceae) based on analysis of whole genome sequences. Int. J. Syst. Evol. Microbiol. 63, 2742–2751. doi: 10.1099/ijs.0.051110-0
PubMed Abstract | CrossRef Full Text | Google Scholar
Urbanczyk, H., Ogura, Y., and Hayashi, T. (2014). Contrasting inter- and intraspecies recombination patterns in the “Harveyi Clade” Vibrio collected over large spatial and temporal scales. Genome Biol. Evol. 7, 71–80. doi: 10.1093/gbe/evu269
PubMed Abstract | CrossRef Full Text | Google Scholar
Urbanczyk, Y., Ogura, Y., Hayashi, T., and Urbanczyk, H. (2016). Genomic evidence that Vibrio inhibens is a heterotypic synonym of Vibrio jasicida. Int. J. Syst. Evol. Microbiol. 66, 3214–3218. doi: 10.1099/ijsem.0.001173
PubMed Abstract | CrossRef Full Text | Google Scholar
Varghese, N. J., Mukherjee, S., Ivanova, N., Konstantinidis, K. T., Mavrommatis, K., Kyrpides, N. C., et al. (2015). Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771. doi: 10.1093/nar/gkv657
PubMed Abstract | CrossRef Full Text | Google Scholar
Yáñez, R., Bastías, R., Higuera, G., Salgado, O., Katharios, P., Romero, J., et al. (2015). Amplification of tlh gene in other Vibrionaceae species by species-specific multiplex PCR of Vibrio parahaemolyticus. Electron. J. Biotechnol. 18, 459–463. doi: 10.1016/j.ejbt.2015.09.007
CrossRef Full Text | Google Scholar
Zerbino, D. R., and Birney, E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829. doi: 10.1101/gr.074492.107
PubMed Abstract | CrossRef Full Text | Google Scholar
Keywords: Vibrio diabolicus, Vibrio antiquarius, Vibrio alginolyticus, deep-sea, taxonomy, genomics, diversity
Citation: Turner JW, Tallman JJ, Macias A, Pinnell LJ, Elledge NC, Nasr Azadani D, Nilsson WB, Paranjpye RN, Armbrust EV and Strom MS (2018) Comparative Genomic Analysis of Vibrio diabolicus and Six Taxonomic Synonyms: A First Look at the Distribution and Diversity of the Expanded Species. Front. Microbiol. 9:1893. doi: 10.3389/fmicb.2018.01893
Received: 11 April 2018; Accepted: 27 July 2018;Published: 15 August 2018.
Edited by:
Frank T. Robb, University of Maryland, Baltimore, United States
Reviewed by:
Charles Lovell, University of South Carolina, United States Iddya Karunasagar, Nitte University, India Christopher John Grim, United States Food and Drug Administration, United States
Copyright © 2018 Turner, Tallman, Macias, Pinnell, Elledge, Nasr Azadani, Nilsson, Paranjpye, Armbrust and Strom. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeffrey W. Turner, amVmZnJleS50dXJuZXJAdGFtdWNjLmVkdQ==
