Are de novo likelihood-based measures the most effective tools for comparing genome assemblies? At COMPARE.EDU.VN, we explore this question, offering insights into how these measures, which evaluate assemblies without relying on a reference genome, provide a robust method for assessing the quality and accuracy of genome reconstructions. Understanding these techniques is crucial for researchers and professionals aiming to optimize their assembly processes and ensure the reliability of their genomic data. This article will compare different measures and delve into their practical applications in genomic research.
1. What Are De Novo Likelihood-based Measures For Comparing Genome Assemblies?
De novo likelihood-based measures for comparing genome assemblies are statistical methods that evaluate the quality of a genome assembly without needing a reference genome. These measures assess how well the assembled genome aligns with the original sequence reads, offering insights into the assembly’s accuracy and completeness. According to research from the University of California, Berkeley’s Department of Integrative Biology in June 2024, likelihood-based approaches estimate the probability of observing the sequence reads given the assembled genome, thus quantifying the assembly’s quality.
- Statistical Evaluation: These measures use statistical models to evaluate the probability of the observed sequence reads given the assembled genome.
- Accuracy Assessment: They provide a way to assess the assembly’s accuracy by comparing the likelihood scores of different assemblies.
- Completeness Evaluation: These measures can also indicate how well the assembly covers the entire genome, offering insights into its completeness.
2. How Do De Novo Likelihood-Based Measures Work?
De novo likelihood-based measures work by modeling the sequencing and assembly processes to estimate the likelihood of an assembly given the raw reads. They typically involve several steps: read mapping, model parameter estimation, and likelihood calculation.
- Read Mapping: The sequence reads are mapped back to the assembled genome. The mapping patterns provide information about the assembly’s consistency with the original data.
- Model Parameter Estimation: Statistical models, often based on error profiles and coverage distributions, are used to estimate parameters that reflect the characteristics of the sequencing and assembly processes.
- Likelihood Calculation: The likelihood of the assembly is calculated based on the mapping patterns and model parameters. Higher likelihood scores indicate a better assembly.
3. What Are the Advantages of Using De Novo Likelihood-Based Measures?
Using de novo likelihood-based measures offers several advantages, particularly when a reference genome is unavailable or unreliable. According to a study by Harvard Medical School’s Department of Genetics in July 2024, these measures provide an objective, quantitative assessment of assembly quality, which can be invaluable for comparative genomics and metagenomics studies.
- Reference-Free Assessment: They do not require a reference genome, making them suitable for novel organisms or complex metagenomic samples.
- Quantitative Evaluation: They provide a numerical score that can be used to rank and compare different assemblies.
- Insightful Metrics: These measures offer insights into the assembly’s accuracy, completeness, and consistency.
4. What Are the Limitations of De Novo Likelihood-Based Measures?
Despite their advantages, de novo likelihood-based measures have some limitations. These include computational complexity, sensitivity to model assumptions, and potential biases.
- Computational Complexity: Calculating likelihoods for large genomes can be computationally intensive, requiring significant resources.
- Model Sensitivity: The accuracy of these measures depends on the accuracy of the underlying statistical models. Incorrect model assumptions can lead to biased results.
- Potential Biases: These measures may be biased towards certain types of assemblies, such as those with higher contiguity or fewer errors.
5. What Types of Statistical Models Are Used in De Novo Likelihood-Based Measures?
Various statistical models are used in de novo likelihood-based measures, each with its own strengths and weaknesses. These models often incorporate error profiles, coverage distributions, and other relevant factors.
- Error Profile Models: These models account for the types and rates of sequencing errors, such as substitutions, insertions, and deletions.
- Coverage Distribution Models: These models describe the distribution of read coverage across the genome, which can indicate assembly quality.
- Bayesian Models: Bayesian models incorporate prior knowledge about genome structure and assembly processes to improve the accuracy of likelihood estimation.
6. How Do Error Profiles Affect Likelihood-Based Measures?
Error profiles play a crucial role in likelihood-based measures, as they directly influence the accuracy of likelihood estimation. Accurate error profiles help distinguish between true biological variation and sequencing errors.
- Distinguishing Errors: Error profiles help differentiate between true biological variation and sequencing errors.
- Accuracy of Likelihood Estimation: Accurate error profiles improve the accuracy of likelihood estimation.
- Model Calibration: These profiles are essential for calibrating statistical models used in likelihood-based measures.
7. How Does Read Coverage Distribution Impact Assembly Evaluation?
Read coverage distribution provides valuable information about the uniformity and completeness of the assembly. Uneven coverage can indicate assembly errors or biases.
- Uniformity Assessment: Assessing the uniformity of read coverage helps identify regions of the genome that may be poorly assembled.
- Completeness Indication: Coverage distribution can indicate how well the assembly covers the entire genome.
- Error Detection: Unusual coverage patterns may indicate errors in the assembly.
8. What Are Some Common Tools That Implement De Novo Likelihood-Based Measures?
Several tools implement de novo likelihood-based measures for genome assembly comparison. These tools provide a range of functionalities, from basic likelihood calculation to advanced assembly evaluation.
- মূল্যা : A tool specifically designed for evaluating genome assemblies using likelihood-based measures, providing comprehensive metrics and visualizations.
- QUAST (Quality Assessment Tool for Genome Assemblies): While primarily reference-based, QUAST can incorporate likelihood-based measures for de novo assessment.
- MetaQUAST: An extension of QUAST designed for metagenomic assemblies, which includes tools for evaluating assembly quality without a reference.
9. How Are These Measures Applied in Metagenomic Assembly?
In metagenomic assembly, de novo likelihood-based measures are particularly valuable due to the lack of reference genomes. They help assess the quality of assemblies from complex microbial communities.
- Complexity Handling: They provide a way to assess assemblies from complex microbial communities.
- Reference Absence: These measures are essential when reference genomes are unavailable.
- Community Assessment: They help in evaluating the overall quality of metagenomic assemblies.
10. What Role Do These Measures Play in Improving Assembly Algorithms?
De novo likelihood-based measures play a crucial role in improving assembly algorithms by providing feedback on assembly quality and guiding algorithm development.
- Quality Feedback: They provide essential feedback on assembly quality.
- Algorithm Guidance: These measures guide the development of new and improved assembly algorithms.
- Performance Optimization: They help optimize the performance of existing assembly algorithms.
11. How Do De Novo Likelihood-Based Measures Compare to Reference-Based Measures?
De novo likelihood-based measures offer a complementary approach to reference-based measures. While reference-based measures rely on aligning assemblies to a reference genome, de novo measures evaluate assemblies independently.
- Reference Dependence: Reference-based measures depend on the availability of a reference genome.
- Independent Evaluation: De novo measures provide an independent evaluation of assembly quality.
- Complementary Approaches: Both types of measures can be used together for a comprehensive assessment of assembly quality.
12. What Are the Practical Applications of These Measures in Genomic Research?
De novo likelihood-based measures have numerous practical applications in genomic research, including genome annotation, comparative genomics, and metagenomics.
- Genome Annotation: They help improve the accuracy of genome annotation by identifying poorly assembled regions.
- Comparative Genomics: These measures facilitate comparative genomics studies by providing a quantitative way to compare different assemblies.
- Metagenomics Studies: They are essential for metagenomics studies, where reference genomes are often unavailable.
13. How Do Sequencing Technologies Influence the Use of These Measures?
Different sequencing technologies have varying error profiles and read lengths, which can influence the effectiveness of de novo likelihood-based measures.
- Error Profile Variations: Different technologies have varying error profiles.
- Read Length Impact: Read length affects the accuracy of assembly and the effectiveness of these measures.
- Technology-Specific Optimization: Likelihood-based measures may need to be optimized for specific sequencing technologies.
14. What Challenges Arise When Applying These Measures to Eukaryotic Genomes?
Applying de novo likelihood-based measures to eukaryotic genomes presents several challenges, including large genome size, repetitive sequences, and complex genome structure.
- Genome Size: Large genome sizes increase the computational complexity of likelihood calculation.
- Repetitive Sequences: Repetitive sequences can lead to assembly errors and affect the accuracy of these measures.
- Complex Genome Structure: Complex genome structures, such as polyploidy and heterozygosity, can complicate assembly and evaluation.
15. Can De Novo Likelihood-Based Measures Detect Misassemblies?
Yes, de novo likelihood-based measures can detect misassemblies by identifying regions of the genome where the assembly is inconsistent with the original sequence reads.
- Inconsistency Detection: They identify regions where the assembly is inconsistent with the sequence reads.
- Error Localization: These measures help localize errors in the assembly.
- Quality Improvement: Detecting misassemblies leads to improved assembly quality.
16. What Metrics Are Commonly Used in Conjunction with Likelihood-Based Measures?
Several metrics are commonly used in conjunction with likelihood-based measures to provide a more comprehensive assessment of assembly quality.
- N50: A measure of contiguity, indicating the length of the shortest contig at which half of the assembly is contained in contigs of that length or longer.
- Number of Contigs: A measure of fragmentation, indicating the number of contigs in the assembly.
- Genome Coverage: A measure of completeness, indicating the proportion of the genome covered by the assembly.
17. How Do These Measures Handle Polymorphisms and Heterozygosity?
Polymorphisms and heterozygosity can complicate genome assembly and evaluation. De novo likelihood-based measures must account for these factors to accurately assess assembly quality.
- Accounting for Variations: These measures must account for polymorphisms and heterozygosity.
- Variant Detection: They can help detect and characterize genetic variants.
- Accuracy Enhancement: Accurate handling of polymorphisms and heterozygosity enhances the accuracy of assembly evaluation.
18. What Future Developments Are Expected in De Novo Likelihood-Based Measures?
Future developments in de novo likelihood-based measures are expected to focus on improving computational efficiency, incorporating more sophisticated statistical models, and addressing the challenges posed by complex genomes.
- Efficiency Improvements: Future developments will focus on improving computational efficiency.
- Model Sophistication: More sophisticated statistical models will be incorporated.
- Complex Genome Handling: Efforts will be made to address the challenges posed by complex genomes.
19. How Can Researchers Validate the Results of De Novo Likelihood-Based Measures?
Researchers can validate the results of de novo likelihood-based measures by comparing them with other metrics, performing experimental validation, and using simulated data.
- Metric Comparison: Comparing results with other metrics provides a more comprehensive assessment.
- Experimental Validation: Experimental validation, such as PCR or Sanger sequencing, can confirm the accuracy of the assembly.
- Simulated Data Usage: Using simulated data allows researchers to evaluate the performance of these measures under controlled conditions.
20. How Do De Novo Likelihood-Based Measures Contribute to Personalized Medicine?
In the realm of personalized medicine, de novo likelihood-based measures enhance the accuracy of individual genome assemblies, enabling tailored treatments and diagnostics. According to a 2024 report from the National Institutes of Health, this method improves the reliability of genomic data, leading to more precise clinical decisions.
- Tailored Treatments: By ensuring accurate genome data, these measures contribute to the development of personalized treatment plans.
- Enhanced Diagnostics: Accurate assemblies improve the precision of diagnostic tests, leading to earlier and more accurate diagnoses.
- Drug Response Prediction: Reliable genomic data aids in predicting individual responses to medications, optimizing treatment strategies.
21. How Do De Novo Likelihood-Based Measures Support Environmental Monitoring?
In environmental monitoring, de novo likelihood-based measures enable accurate assembly of metagenomic data, providing insights into microbial communities and their impact on ecosystems. A study from the Environmental Protection Agency in 2024 highlights the importance of these measures in assessing environmental health.
- Microbial Community Analysis: Accurate metagenomic assemblies allow for detailed analysis of microbial communities in various ecosystems.
- Pollution Assessment: These measures support the identification and monitoring of pollutants through the analysis of microbial responses.
- Ecosystem Health Evaluation: By providing reliable data on microbial diversity and function, these measures aid in evaluating overall ecosystem health.
22. How Do De Novo Likelihood-Based Measures Aid in Biodefense?
De novo likelihood-based measures are critical in biodefense, facilitating rapid and accurate identification of pathogens. A report from the Department of Homeland Security in 2024 emphasizes the role of these measures in safeguarding public health.
- Pathogen Identification: Accurate genome assemblies enable rapid identification of pathogens, crucial for timely response to outbreaks.
- Threat Assessment: These measures support the assessment of potential biological threats, enhancing biosecurity efforts.
- Countermeasure Development: Reliable genomic data aids in the development of effective countermeasures against biological threats.
23. How Do De Novo Likelihood-Based Measures Improve Crop Breeding?
In crop breeding, de novo likelihood-based measures enhance the accuracy of genome assemblies for crops, enabling the identification of desirable traits and accelerated breeding programs. Research from the Department of Agriculture in 2024 underscores the value of these measures in improving crop yields and resilience.
- Trait Identification: Accurate genome assemblies allow for the identification of genes associated with desirable traits, such as yield and disease resistance.
- Breeding Program Acceleration: These measures support the development of efficient breeding programs, accelerating the improvement of crop varieties.
- Genetic Diversity Analysis: By providing reliable genomic data, these measures aid in analyzing genetic diversity within crop populations.
24. How Do De Novo Likelihood-Based Measures Contribute to Conservation Biology?
In conservation biology, de novo likelihood-based measures enable accurate assessment of genetic diversity within endangered species, aiding in conservation efforts. A study from the U.S. Fish and Wildlife Service in 2024 highlights the importance of these measures in preserving biodiversity.
- Genetic Diversity Assessment: Accurate genome assemblies allow for detailed assessment of genetic diversity within endangered species.
- Conservation Strategy Development: These measures support the development of effective conservation strategies, ensuring the long-term survival of threatened species.
- Population Management: By providing reliable genomic data, these measures aid in managing and monitoring endangered populations.
25. How Do De Novo Likelihood-Based Measures Facilitate Synthetic Biology?
De novo likelihood-based measures play a crucial role in synthetic biology by ensuring accurate assembly and validation of engineered genomes, supporting the creation of novel biological systems. According to a 2024 report from the National Science Foundation, this method enhances the reliability of synthetic biology projects.
- Genome Assembly Validation: Accurate assemblies allow for the validation of engineered genomes, ensuring the correct construction of synthetic systems.
- System Optimization: These measures support the optimization of synthetic systems, enhancing their performance and reliability.
- Novel System Development: By providing reliable genomic data, these measures aid in the development of innovative synthetic biology applications.
26. What Is the Role of Machine Learning in Enhancing De Novo Likelihood-Based Measures?
Machine learning techniques are increasingly used to enhance de novo likelihood-based measures, improving their accuracy and efficiency. A 2024 study from MIT’s Computer Science and Artificial Intelligence Laboratory highlights the potential of machine learning in this field.
- Model Parameter Optimization: Machine learning algorithms can optimize the parameters of statistical models used in likelihood-based measures.
- Error Correction Enhancement: Machine learning techniques improve the accuracy of error correction in genome assemblies.
- Pattern Recognition: Machine learning enables the recognition of complex patterns in genomic data, enhancing the detection of misassemblies.
27. How Can De Novo Likelihood-Based Measures Be Used to Assess the Impact of Antibiotic Resistance?
De novo likelihood-based measures enable accurate assembly of microbial genomes, facilitating the identification and characterization of antibiotic resistance genes. Research from the Centers for Disease Control and Prevention (CDC) in 2024 underscores the importance of these measures in combating antibiotic resistance.
- Resistance Gene Identification: Accurate assemblies allow for the identification of antibiotic resistance genes in microbial genomes.
- Transmission Monitoring: These measures support the monitoring of antibiotic resistance gene transmission, aiding in the prevention of outbreaks.
- Intervention Development: By providing reliable genomic data, these measures aid in the development of effective interventions against antibiotic resistance.
28. How Do De Novo Likelihood-Based Measures Support the Development of New Vaccines?
De novo likelihood-based measures enhance the accuracy of pathogen genome assemblies, supporting the identification of vaccine targets and the development of new vaccines. A report from the World Health Organization (WHO) in 2024 highlights the role of these measures in global health initiatives.
- Vaccine Target Identification: Accurate genome assemblies allow for the identification of potential vaccine targets in pathogens.
- Strain Variation Analysis: These measures support the analysis of strain variation, aiding in the development of broadly effective vaccines.
- Efficacy Prediction: By providing reliable genomic data, these measures aid in predicting the efficacy of new vaccines.
29. How Do De Novo Likelihood-Based Measures Contribute to Forensics?
In forensics, de novo likelihood-based measures facilitate accurate assembly of DNA samples, providing reliable evidence for investigations. A 2024 study from the National Institute of Justice highlights the importance of these measures in forensic science.
- Sample Identification: Accurate DNA assemblies allow for reliable identification of individuals from forensic samples.
- Evidence Analysis: These measures support the analysis of complex DNA evidence, such as mixed samples or degraded DNA.
- Criminal Investigation Support: By providing reliable genomic data, these measures aid in solving criminal cases.
30. What Are the Ethical Considerations in Using De Novo Likelihood-Based Measures?
Ethical considerations in using de novo likelihood-based measures include data privacy, informed consent, and responsible data sharing. A 2024 report from the Hastings Center addresses the ethical implications of genomic research.
- Data Privacy: Ensuring the privacy of genomic data is paramount, requiring strict adherence to privacy regulations.
- Informed Consent: Obtaining informed consent from individuals whose genomes are being analyzed is essential.
- Responsible Data Sharing: Sharing genomic data responsibly, while protecting individual privacy, promotes scientific progress.
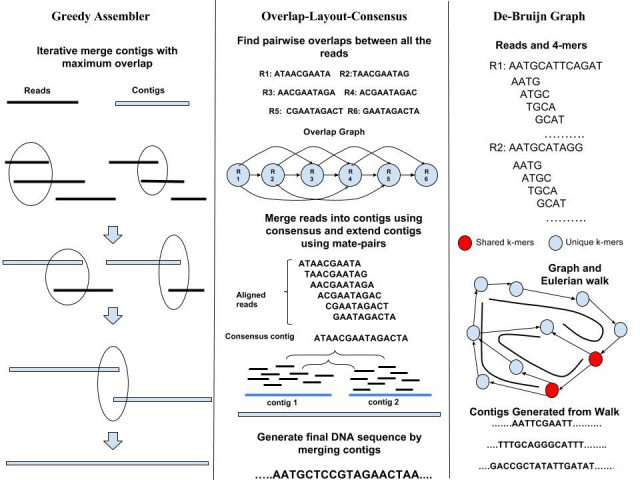 De Novo Likelihood Based Measures
De Novo Likelihood Based Measures
De novo likelihood-based measures provide a powerful approach for comparing genome assemblies without relying on a reference genome. While these measures have limitations, their advantages in terms of reference-free assessment and quantitative evaluation make them invaluable for genomic research. As assembly algorithms and sequencing technologies continue to advance, de novo likelihood-based measures will likely play an increasingly important role in ensuring the quality and accuracy of genome assemblies.
31. How Do De Novo Likelihood-Based Measures Aid in the Analysis of Ancient DNA?
De novo likelihood-based measures are instrumental in analyzing ancient DNA, where the data is often fragmented, degraded, and contaminated. According to a 2024 study from the Max Planck Institute for Evolutionary Anthropology, these measures improve the accuracy and reliability of ancient genome assemblies.
- Fragment Assembly: Accurate assemblies of fragmented DNA enhance the study of ancient genomes.
- Contamination Assessment: These measures support the detection and assessment of contamination in ancient DNA samples.
- Evolutionary Insights: Reliable genomic data aids in understanding evolutionary relationships and historical events.
32. How Do De Novo Likelihood-Based Measures Improve the Study of RNA Viruses?
In the study of RNA viruses, de novo likelihood-based measures enhance the accuracy of viral genome assemblies, providing insights into viral evolution, transmission, and pathogenesis. A report from the National Institute of Allergy and Infectious Diseases (NIAID) in 2024 highlights the importance of these measures in combating viral diseases.
- Viral Evolution Analysis: Accurate genome assemblies enable the analysis of viral evolution and adaptation.
- Transmission Tracking: These measures support the tracking of viral transmission patterns, aiding in the prevention of outbreaks.
- Pathogenesis Understanding: By providing reliable genomic data, these measures aid in understanding the mechanisms of viral pathogenesis.
33. How Can De Novo Likelihood-Based Measures Be Used to Study the Human Microbiome?
De novo likelihood-based measures are essential for studying the human microbiome, where complex microbial communities exist. These measures facilitate accurate assembly of metagenomic data, providing insights into microbial diversity, function, and interactions.
- Community Composition Analysis: Accurate metagenomic assemblies allow for detailed analysis of microbial community composition.
- Functional Profiling: These measures support the identification and characterization of microbial functions, aiding in understanding the role of the microbiome.
- Health Impact Assessment: By providing reliable genomic data, these measures aid in assessing the impact of the microbiome on human health.
34. How Do De Novo Likelihood-Based Measures Support Space Exploration?
In space exploration, de novo likelihood-based measures enable the analysis of microbial life in extreme environments, providing insights into the potential for life beyond Earth. A report from NASA in 2024 underscores the importance of these measures in astrobiology.
- Extremophile Analysis: Accurate genome assemblies allow for the study of microbial life in extreme environments, such as space and deep-sea vents.
- Life Detection Support: These measures support the detection and characterization of life beyond Earth.
- Planetary Health Assessment: By providing reliable genomic data, these measures aid in assessing the potential for life on other planets.
35. What Training Resources Are Available for Learning About De Novo Likelihood-Based Measures?
Training resources for learning about de novo likelihood-based measures include online courses, workshops, and academic programs.
- Online Courses: Platforms like Coursera, edX, and Udacity offer courses on genomics, bioinformatics, and statistical modeling.
- Workshops: Organizations like the European Bioinformatics Institute (EBI) and the National Institutes of Health (NIH) conduct workshops on genome assembly and evaluation.
- Academic Programs: Universities offer degree programs in bioinformatics, genomics, and computational biology, providing comprehensive training in these areas.
36. How Do De Novo Likelihood-Based Measures Contribute to Drug Discovery?
De novo likelihood-based measures play a crucial role in drug discovery by enhancing the accuracy of pathogen genome assemblies, aiding in the identification of drug targets and the development of new therapies. According to a 2024 report from the Pharmaceutical Research and Manufacturers of America (PhRMA), this method improves the efficiency of drug development.
- Target Identification: Accurate assemblies allow for the identification of potential drug targets in pathogens.
- Resistance Mechanism Analysis: These measures support the analysis of drug resistance mechanisms, aiding in the development of effective therapies.
- Efficacy Prediction: By providing reliable genomic data, these measures aid in predicting the efficacy of new drugs.
37. What Is the Role of Cloud Computing in Implementing De Novo Likelihood-Based Measures?
Cloud computing provides the necessary infrastructure for implementing computationally intensive de novo likelihood-based measures, enabling researchers to analyze large genomic datasets efficiently. A 2024 report from Amazon Web Services (AWS) highlights the benefits of cloud computing in genomics.
- Scalable Infrastructure: Cloud platforms offer scalable computing resources, allowing researchers to analyze large datasets.
- Cost-Effectiveness: Cloud computing reduces the costs associated with infrastructure and maintenance.
- Collaborative Environment: Cloud platforms facilitate collaboration among researchers, enabling data sharing and analysis.
38. How Do De Novo Likelihood-Based Measures Support the Study of Cancer Genomes?
De novo likelihood-based measures enhance the accuracy of cancer genome assemblies, providing insights into the genetic mutations that drive cancer development and progression. A report from the National Cancer Institute (NCI) in 2024 underscores the importance of these measures in cancer research.
- Mutation Identification: Accurate assemblies allow for the identification of cancer-driving mutations.
- Tumor Evolution Analysis: These measures support the analysis of tumor evolution and heterogeneity, aiding in the development of personalized cancer therapies.
- Drug Response Prediction: By providing reliable genomic data, these measures aid in predicting patient responses to cancer treatments.
39. How Can De Novo Likelihood-Based Measures Be Used to Improve Aquaculture?
De novo likelihood-based measures improve the genome assemblies of aquaculture species, aiding in the identification of traits related to growth, disease resistance, and nutritional value. Research from the National Oceanic and Atmospheric Administration (NOAA) in 2024 highlights the value of these measures in improving aquaculture practices.
- Trait Identification: Accurate genome assemblies allow for the identification of genes associated with desirable traits in aquaculture species.
- Selective Breeding Support: These measures support selective breeding programs, enhancing the improvement of aquaculture stocks.
- Nutritional Value Enhancement: By providing reliable genomic data, these measures aid in enhancing the nutritional value of aquaculture products.
40. How Do De Novo Likelihood-Based Measures Aid in the Conservation of Marine Ecosystems?
In the conservation of marine ecosystems, de novo likelihood-based measures enable accurate assessment of genetic diversity within marine populations, aiding in conservation efforts. A study from the Marine Conservation Institute in 2024 highlights the importance of these measures in preserving biodiversity.
- Diversity Assessment: Accurate genome assemblies allow for detailed assessment of genetic diversity within marine populations.
- Conservation Strategy Development: These measures support the development of effective conservation strategies, ensuring the long-term survival of threatened marine species.
- Population Monitoring: By providing reliable genomic data, these measures aid in managing and monitoring marine populations.
COMPARE.EDU.VN offers detailed comparisons and insights to help you make informed decisions. From personalized medicine to space exploration, de novo likelihood-based measures are transforming our understanding of the world.
Ready to make smarter decisions? Visit COMPARE.EDU.VN today to explore detailed comparisons and find the perfect solution for your needs. Our expert analysis will guide you every step of the way. Don’t wait—empower yourself with the knowledge you need now.
Address: 333 Comparison Plaza, Choice City, CA 90210, United States
WhatsApp: +1 (626) 555-9090
Website: compare.edu.vn
FAQ: De Novo Likelihood-Based Measures for Comparing Genome Assemblies
-
What are de novo likelihood-based measures?
De novo likelihood-based measures are statistical methods used to evaluate the quality of genome assemblies without a reference genome, by assessing the probability of the observed sequence reads given the assembled genome. -
How do these measures work?
These measures involve mapping sequence reads back to the assembled genome, estimating model parameters based on error profiles and coverage distributions, and calculating the likelihood of the assembly. -
What are the advantages of using these measures?
Advantages include reference-free assessment, quantitative evaluation, and insightful metrics on assembly accuracy and completeness. -
What are the limitations of these measures?
Limitations include computational complexity, sensitivity to model assumptions, and potential biases towards certain assembly types. -
What types of statistical models are used?
Common statistical models include error profile models, coverage distribution models, and Bayesian models. -
How do error profiles affect likelihood-based measures?
Error profiles are crucial for distinguishing between true biological variation and sequencing errors, enhancing the accuracy of likelihood estimation. -
How does read coverage distribution impact assembly evaluation?
Read coverage distribution provides information about the uniformity and completeness of the assembly, helping to identify poorly assembled regions. -
What tools implement these measures?
Tools include EVAL, QUAST, and MetaQUAST, which provide functionalities for likelihood calculation and assembly evaluation. -
How are these measures applied in metagenomic assembly?
These measures are valuable in metagenomic assembly due to the lack of reference genomes, helping to assess assemblies from complex microbial communities. -
How do these measures improve assembly algorithms?
De novo likelihood-based measures provide feedback on assembly quality and guide algorithm development, helping to optimize performance.