Are you struggling to understand the complex effects of environmental chemicals on human health? COMPARE.EDU.VN provides a powerful solution: the Comparative Toxicogenomics Database (CTD). This curated database connects chemicals, genes, and diseases, enabling users to uncover hidden relationships and formulate testable hypotheses about environmental diseases. Explore how CTD functions as a knowledge base and discovery tool to unlock insights into toxicogenomics, chemical-gene interactions, and environmental health.
1. What is a Comparative Toxicogenomics Database?
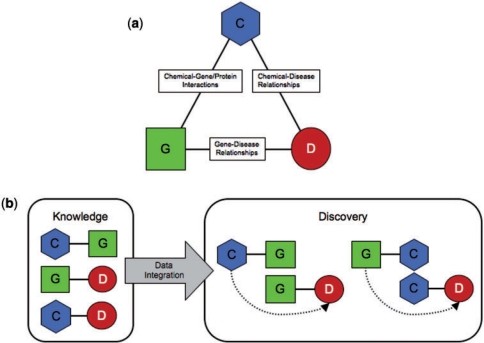
A comparative toxicogenomics database is a specialized resource that integrates data about chemicals, genes, and diseases to help understand how environmental agents affect human health. It combines manually curated information with computational tools to reveal connections and generate hypotheses about the mechanisms behind environmental diseases. The Comparative Toxicogenomics Database (CTD) is a prime example of such a database.
1.1. Key Components of a Comparative Toxicogenomics Database
- Chemical Data: Information on environmental chemicals, including names, synonyms, structures, and identifiers.
- Gene Data: Official gene symbols, names, sequences, and links to other gene databases.
- Disease Data: Disease terms from controlled vocabularies like MeSH and OMIM, along with associated information.
- Interaction Data: Curated relationships between chemicals and genes, chemicals and diseases, and genes and diseases.
- Integration Tools: Tools for querying, analyzing, and visualizing the integrated data to discover new connections.
1.2. How Does CTD Stand Out Among Other Databases?
The Comparative Toxicogenomics Database (CTD) distinguishes itself through several unique features:
- Focus on Environmental Chemicals: CTD specifically emphasizes environmental chemicals, differentiating it from databases primarily focused on drugs or other small molecules.
- Manual Curation: Professional biocurators manually extract and validate chemical-gene interactions, chemical-disease relationships, and gene-disease relationships from scientific literature. This ensures high accuracy and reliability.
- Controlled Vocabularies: CTD uses controlled vocabularies and hierarchies for chemicals, genes, and diseases.
- Integration of Data: CTD integrates curated and imported data to allow users to explore the connections between chemicals, genes, and diseases comprehensively.
- Discovery Tool: In addition to being a repository for information, CTD serves as a resource for generating novel hypotheses about environmental diseases and chemical actions.
1.3. Who Benefits from Using a Comparative Toxicogenomics Database?
- Researchers: Can use CTD to generate hypotheses and design experiments.
- Toxicologists: Can study the mechanisms of chemical toxicity.
- Environmental Health Scientists: Can investigate the impact of environmental exposures on human health.
- Clinicians: Can understand the genetic and environmental factors influencing disease susceptibility.
2. Why is a Comparative Toxicogenomics Database Important?
A comparative toxicogenomics database is vital because it helps bridge the gap between environmental exposures and human health outcomes. By integrating data from diverse sources, it allows researchers and healthcare professionals to understand the complex interactions between chemicals, genes, and diseases.
2.1. Understanding Environmental Chemical Effects
Environmental chemicals play a significant role in the development of many human diseases. A comparative toxicogenomics database provides a structured way to understand how these chemicals interact with genes and proteins, influencing biological processes and disease development.
2.2. Generating Testable Hypotheses
One of the primary benefits of a comparative toxicogenomics database is its ability to generate testable hypotheses. By identifying connections between chemicals, genes, and diseases, researchers can formulate specific questions and design experiments to validate these relationships.
2.3. Supporting Evidence-Based Decisions
The information within a comparative toxicogenomics database supports evidence-based decisions in public health and environmental policy. By providing a comprehensive view of chemical-gene-disease interactions, policymakers can make informed decisions to protect public health.
2.4. Facilitating Collaboration
A comparative toxicogenomics database acts as a central resource that facilitates collaboration among researchers, clinicians, and policymakers. By providing a common platform for sharing data and insights, it promotes interdisciplinary collaboration and accelerates the pace of scientific discovery.
2.5. Providing a Foundation for Personalized Medicine
The study of toxicogenomics also contributes to the progress of customized medicine by assisting in the discovery of how genetic variations affect people’s reactions to environmental variables. This insight makes it easier to customize care strategies and preventative actions based on a person’s genetic make-up and exposure background.
3. How Does the Comparative Toxicogenomics Database Work?
The Comparative Toxicogenomics Database (CTD) operates by manually curating data from scientific literature and integrating it using controlled vocabularies. This process involves several key steps to ensure accuracy and consistency.
3.1. Manual Curation Process
Professional biocurators read scientific articles to identify and extract three types of core data:
- Chemical-Gene Interactions: How chemicals interact with genes or proteins.
- Chemical-Disease Relationships: Direct relationships between chemicals and diseases.
- Gene-Disease Relationships: Associations between genes and diseases.
3.2. Use of Controlled Vocabularies
CTD uses controlled vocabularies to standardize the data:
- Chemicals: Terms from the Medical Subject Headings (MeSH) vocabulary.
- Genes: Official gene symbols and names from the National Center for Biotechnology Information’s (NCBI) Entrez-Gene database.
- Diseases: Terms from the disease subset of MeSH and OMIM.
3.3. Data Integration and Inference
The curated data are integrated to create a triad of chemicals, genes, and diseases. This integration allows CTD to infer novel connections:
- If chemical A interacts with gene B, and gene B is associated with disease C, then CTD infers that chemical A may have a relationship with disease C.
3.4. Querying and Data Retrieval
Users can query CTD using a keyword search box or detailed query pages. Search parameters include:
- Chemicals
- Genes
- Diseases
- GO terms
- Organisms
- References
3.5. Data Visualization and Analysis
CTD provides various ways to view and analyze data:
- Basic Information Pages: Provide names, synonyms, identifiers, and links to external resources.
- Interaction Pages: List curated chemical-gene interactions.
- Chemical/Gene/Disease Pages: List associated chemicals, genes, or diseases, along with direct or inferred associations.
4. What Information Can You Find in the Comparative Toxicogenomics Database?
The Comparative Toxicogenomics Database (CTD) contains a wealth of information organized around chemicals, genes, and diseases. Understanding the types of data available can help users effectively leverage the database for their research.
4.1. Chemical Data
- Names and Synonyms: Comprehensive list of names and synonyms for each chemical.
- Chemical Structures: Chemical structures and identifiers (e.g., CAS registry numbers).
- Interactions: List of genes that interact with the chemical, along with detailed interaction types.
- Associated Diseases: Diseases associated with the chemical, including direct and inferred relationships.
4.2. Gene Data
- Official Symbols and Names: Official gene symbols and names from NCBI Entrez-Gene.
- Sequences: Protein and nucleic acid sequences.
- Interactions: List of chemicals that interact with the gene, along with detailed interaction types.
- Associated Diseases: Diseases associated with the gene, including direct and inferred relationships.
- GO Annotations: Gene Ontology (GO) annotations providing information on gene function and localization.
4.3. Disease Data
- Disease Terms: Standardized disease terms from MeSH and OMIM.
- Associated Genes: Genes associated with the disease, including direct and inferred relationships.
- Associated Chemicals: Chemicals associated with the disease, including direct and inferred relationships.
4.4. Interaction Data
- Chemical-Gene Interactions: Detailed information on how chemicals interact with genes, including the type of interaction (e.g., binding, phosphorylation, activity).
- Organism: The organism in which the interaction was observed.
- Reference: Link to the PubMed identifier (PMID) of the scientific article supporting the interaction.
4.5. Inferred Relationships
- Inferred Chemical-Disease Relationships: Potential relationships between chemicals and diseases inferred through gene interactions.
- Inferred Gene-Disease Relationships: Potential relationships between genes and diseases inferred through chemical interactions.
5. How to Use the Comparative Toxicogenomics Database for Research?
Using the Comparative Toxicogenomics Database (CTD) effectively can significantly enhance research in toxicology, environmental health, and related fields. Here are some practical steps and strategies to maximize the database’s utility.
5.1. Formulating a Research Question
Start by defining a clear research question. For example:
- “What are the potential environmental chemicals that may contribute to autism?”
- “Which genes are affected by exposure to bisphenol A (BPA)?”
- “What are the molecular mechanisms linking arsenic exposure to lung cancer?”
5.2. Querying the Database
Use the CTD’s query tools to retrieve relevant data. You can use the keyword search box or the detailed query pages.
- Keyword Search: Enter specific terms (e.g., “bisphenol A,” “autism,” “arsenic”) into the search box.
- Detailed Query: Use the Gene, Interaction, or Reference Query pages for more specific searches.
5.3. Navigating Search Results
CTD organizes data on tabbed pages:
- Basic Information: Provides names, synonyms, identifiers, and links to external resources.
- Interactions: Lists curated chemical-gene interactions.
- Genes/Chemicals/Diseases: Lists associated chemicals, genes, or diseases, along with direct or inferred associations.
5.4. Analyzing and Interpreting Data
- Identify Key Interactions: Look for interactions that are well-supported by multiple studies.
- Explore Inferred Relationships: Use inferred relationships to generate new hypotheses.
- Consider Organism Specificity: Pay attention to the organism in which the interaction was observed.
- Evaluate References: Review the original scientific articles to understand the context and experimental details of the curated data.
5.5. Generating Hypotheses
Use the data from CTD to generate testable hypotheses. For example:
- “Bisphenol A exposure may affect the expression of gene X, leading to disease Y.”
- “Chemical A may interact with gene B in a way that contributes to the development of disease C.”
5.6. Validating Hypotheses
Design experiments to validate the hypotheses generated from CTD data. This may involve:
- In vitro studies to examine chemical-gene interactions.
- In vivo studies to assess the effects of chemical exposure on gene expression and disease development.
- Epidemiological studies to investigate the association between chemical exposure and disease incidence in human populations.
6. What are the Limitations of the Comparative Toxicogenomics Database?
While the Comparative Toxicogenomics Database (CTD) is a valuable resource, it is important to be aware of its limitations to use it effectively and interpret its data accurately.
6.1. Curation Bias
- Limited Scope: CTD primarily focuses on environmental chemicals, which may exclude relevant information about other types of chemicals or agents.
- Publication Bias: The database relies on published literature, which may be subject to publication bias (i.e., studies with positive or significant results are more likely to be published).
- Language Bias: Curation is typically limited to English-language publications, potentially missing relevant data from studies published in other languages.
6.2. Data Accuracy and Completeness
- Manual Curation Limitations: Although manual curation ensures high accuracy, it is a time-consuming process, and the database may not always be up-to-date with the latest research.
- Incomplete Data: Not all chemical-gene-disease relationships have been fully elucidated, and the database may contain incomplete or missing information.
- Data Conflicts: Different studies may report conflicting results, which can be challenging to reconcile.
6.3. Inference Limitations
- Inference Caveats: Inferred relationships are based on indirect connections and may not always be accurate or biologically relevant.
- Lack of Experimental Validation: Inferred relationships require experimental validation to confirm their validity.
6.4. Vocabulary Limitations
- Vocabulary Scope: The controlled vocabularies used in CTD may not cover all possible terms or concepts, potentially limiting the scope of the data.
- Vocabulary Updates: Vocabularies may not be updated as frequently as needed to reflect new discoveries or changes in terminology.
6.5. Technical Limitations
- Query Limitations: Complex queries may be difficult to formulate, and the search functionality may not always retrieve all relevant data.
- Data Download Limitations: Downloading large datasets may be cumbersome or time-consuming.
7. Future Directions for Comparative Toxicogenomics Databases
The field of comparative toxicogenomics is continually evolving, and there are several exciting directions for future development of databases like CTD.
7.1. Enhancing Data Curation
- Text Mining Integration: Incorporating text mining tools to automate the initial screening of literature and improve the efficiency of manual curation.
- Expanding Data Sources: Including data from a wider range of sources, such as grey literature, unpublished studies, and proprietary datasets.
- Community Curation: Engaging the scientific community in data curation through collaborative projects and citizen science initiatives.
7.2. Improving Data Integration
- Standardized Data Formats: Adopting standardized data formats and ontologies to facilitate data sharing and integration across different databases.
- Integrating “Omics” Data: Incorporating data from genomics, transcriptomics, proteomics, and metabolomics studies to provide a more comprehensive view of chemical-gene-disease relationships.
- Network Analysis Tools: Developing advanced network analysis tools to identify complex interactions and pathways.
7.3. Enhancing Data Visualization
- Interactive Visualizations: Creating interactive visualizations to allow users to explore data in a more intuitive and engaging way.
- Customizable Dashboards: Developing customizable dashboards to allow users to track and monitor specific chemicals, genes, or diseases of interest.
7.4. Addressing Ethical and Social Issues
- Data Privacy: Ensuring the privacy and confidentiality of sensitive data, such as individual-level health information.
- Data Equity: Promoting equitable access to data and resources, particularly for researchers in low-resource settings.
- Transparency: Being transparent about the limitations of the data and the methods used to generate it.
8. Examples of Successful Applications of the Comparative Toxicogenomics Database
The Comparative Toxicogenomics Database (CTD) has been successfully applied in various research areas, demonstrating its value in advancing our understanding of environmental health.
8.1. Predicting Disease Associations
- Arsenic and Disease: CTD was used to analyze the arsenic data set, correctly predicting types of diseases that may be associated with arsenic exposure and identifying genes involved in modulating arsenic-related diseases, such as lung cancer and diabetes.
- Autism and Environmental Exposure: CTD has been used to identify potential environmental chemicals that may contribute to autism by examining chemicals that interact with genes known to be associated with autism.
8.2. Understanding Molecular Mechanisms
- Bisphenol A (BPA) and Gene Interactions: Researchers have used CTD to explore the molecular mechanisms by which BPA interacts with genes, providing insights into the potential health effects of BPA exposure.
- Heavy Metals and Limb Development: CTD has been used to identify proteins involved in limb development that interact with heavy metals, helping to understand the developmental toxicity of these chemicals.
8.3. Identifying Therapeutic Targets
- Chemical-Gene-Disease Networks: CTD can be used to identify potential therapeutic targets by examining chemical-gene-disease networks. For example, by identifying genes that are affected by a chemical and are also associated with a disease, researchers can pinpoint potential targets for drug development.
8.4. Supporting Risk Assessment
- Environmental Chemicals and Health Risks: CTD provides data that can be used to assess the potential health risks associated with exposure to environmental chemicals. By integrating data on chemical-gene interactions and disease associations, CTD helps to identify chemicals that pose the greatest risk to human health.
8.5. Facilitating Data Integration
- Cross-Database Integration: CTD enhances its core data pages with links to external resources, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) and the Gene Ontology (GO), facilitating data integration and providing a more comprehensive view of chemical-gene-disease relationships.
9. How Does COMPARE.EDU.VN Utilize Comparative Toxicogenomics Database?
COMPARE.EDU.VN leverages the power of the Comparative Toxicogenomics Database (CTD) to provide users with comprehensive and reliable comparisons of products, services, and ideas related to environmental health.
9.1. Providing Detailed Chemical Information
COMPARE.EDU.VN uses CTD data to provide detailed information on environmental chemicals, including their names, synonyms, structures, and interactions with genes and diseases. This information helps users understand the potential health effects of different chemicals and make informed decisions about their exposure.
9.2. Offering Gene-Environment Interaction Analysis
By integrating CTD data, COMPARE.EDU.VN offers gene-environment interaction analysis, allowing users to explore how genetic factors may influence an individual’s susceptibility to environmental exposures. This analysis helps users understand the complex interplay between genes and the environment in determining health outcomes.
9.3. Supporting Informed Decision-Making
COMPARE.EDU.VN utilizes CTD data to support informed decision-making by providing users with evidence-based comparisons of products and services related to environmental health. This ensures that users have access to the most accurate and up-to-date information when making decisions about their health and well-being.
9.4. Connecting to External Resources
COMPARE.EDU.VN enhances its content with links to external resources, such as KEGG and GO, allowing users to explore data in more detail and gain a more comprehensive understanding of chemical-gene-disease relationships.
9.5. Promoting Transparency and Accuracy
COMPARE.EDU.VN promotes transparency and accuracy by relying on CTD’s manually curated data and controlled vocabularies. This ensures that the information provided is reliable and consistent, allowing users to trust the comparisons and make informed decisions with confidence.
10. Frequently Asked Questions (FAQ) About Comparative Toxicogenomics Database
Here are some frequently asked questions about comparative toxicogenomics databases to help you better understand this valuable resource.
10.1. What is the main purpose of a comparative toxicogenomics database?
The main purpose is to integrate data about chemicals, genes, and diseases to understand how environmental agents affect human health.
10.2. How is data curated in the Comparative Toxicogenomics Database?
Professional biocurators manually extract and validate data from scientific literature.
10.3. What types of data are included in the Comparative Toxicogenomics Database?
Data include chemical-gene interactions, chemical-disease relationships, and gene-disease relationships.
10.4. How can I query the Comparative Toxicogenomics Database?
You can use the keyword search box or detailed query pages to search for specific chemicals, genes, or diseases.
10.5. What are inferred relationships in the Comparative Toxicogenomics Database?
Inferred relationships are potential connections between chemicals, genes, and diseases based on indirect interactions.
10.6. Can the Comparative Toxicogenomics Database help generate hypotheses?
Yes, by identifying connections between chemicals, genes, and diseases, CTD can help researchers generate testable hypotheses.
10.7. Are there any limitations to using the Comparative Toxicogenomics Database?
Limitations include curation bias, data accuracy, inference limitations, and vocabulary limitations.
10.8. How is COMPARE.EDU.VN using the Comparative Toxicogenomics Database?
COMPARE.EDU.VN uses CTD data to provide detailed chemical information, gene-environment interaction analysis, and support informed decision-making.
10.9. How often is the Comparative Toxicogenomics Database updated?
The database is updated monthly to incorporate the latest research findings.
10.10. Where can I find the Comparative Toxicogenomics Database?
You can access the Comparative Toxicogenomics Database at http://ctd.mdibl.org.
Unlock the power of comparative analysis! Visit COMPARE.EDU.VN today to discover detailed comparisons and make informed decisions. Our expert insights will help you navigate complex choices with confidence. Don’t make a decision without us!
Address: 333 Comparison Plaza, Choice City, CA 90210, United States
Whatsapp: +1 (626) 555-9090
Website: compare.edu.vn